













| General Information: |
|
| Name(s) found: |
gi|42566961
[NCBI NR]
gi|28973777 [NCBI NR] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 558 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein folding
[IEA][ISS]
|
| Molecular Function: |
heat shock protein binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESNKEEAKR ALDIAEKKLS KNDYNRAKRY AKKAHRMYPN LVGLEQVLIM IDVYISATNK 60
61 INGEADWYRV LGVDPLADDE AVKKRYRKLA LLLHPDKNRF TGAEGAFKLI LEAWDLLSDK 120
121 SQRSSYDQKR KSNQVKQRTS GMQKPKRSST PKPTESDKPA SSYGPTPPPE PRPKRRPRPN 180
181 IPEPDIPMPM PTRHKPKSKP DISLTTVKVG TFWTVCNRCK TYCEFMRASC LNKTVPCPNC 240
241 GKYFIATVIP SELVNGRLVI RLSPCNQSTW KKASDDTSST SAMAKTVPER VKRWFDPMPE 300
301 MDSSTQKIGT FWTVCSRCKT HCKLVRANHL NKTFPCPNCS QEFVAAEMII EVINGGPVIK 360
361 LSPSVQSNSK RTRDASSTTS ASDKAQRTAY QKEEVLKREK IDPDVASAAT GENVKVAETL 420
421 FKEPMTTGNE NSTTEEGIVK ESETLCKEPM TTGNENSTTE EGNVKEAETF FEEPMTTGNV 480
481 KEVETLFEEP MTTDNENSTP EAERLFKKPM STGDENSTHE AQRLFKKPMT TGNANSNLEA 540
541 QRCFLRIWQR EMQSLLLK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
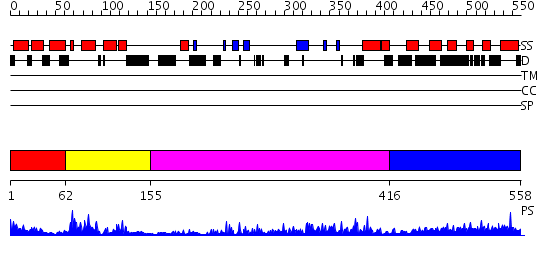
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..61] | 4.09691 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 2 | View Details | [62..154] | 22.522879 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 3 | View Details | [155..415] | 35.0 | The crystal structure of Hsp40 Ydj1 | |
| 4 | View Details | [416..558] | 1.17 | No description for 2j63A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)