













| General Information: |
|
| Name(s) found: |
INVH_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 633 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
catalytic activity
[IEA]
beta-fructofuranosidase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNAITFLGNS TMIPSQCILR AFTRISPSKY IRDTSFRSYP SRFSSCINQY RNADSDRIIR 60
61 PTNAVPFCTD RQSSVTAQVV SEARSHSAST TCANDTTLDQ IYTKNGLNVK PLVVERLKRD 120
121 EKDEEAVNED EEGVKRDGFE GVKCNDVEEE AWRLLRDSIV TYCDSPVGTV AAKDPTDTTP 180
181 SNYDQVFIRD FVPSALAFLL KGESEIVRNF LLHTLQLQSW EKTVDCYSPG QGLMPASFKV 240
241 RTLPLEEDKF EEVLDPDFGE AAIGRVAPVD SGLWWIILLR AYGKITGDYS LQERIDVQTG 300
301 IKMIANLCLA DGFDMFPTLL VTDGSCMIDR RMGIHGHPLE IQALFYSALR SSREMITVND 360
361 SSKNIIKTIS NRLSALSFHI RENYWVDKNK INEIYRYKTE EYSMDATNKF NIYPEQVSPW 420
421 LMDWVPESPD SGFLIGNLQP AHMDFRFFTL GNLWSIISSL GTPKQNQAIL NLVEEKWDDL 480
481 VGHMPLKICY PALESSEWHI ITGSDPKNTP WSYHNGGSWP TLLWQFTLAC IKMGRPELAE 540
541 KAVTLAEKRL QADRWPEYYD TRDGKFIGKQ SRLYQTWTIA GFLTSKQLLQ NPEIASSLFW 600
601 EEDLELLESC VCVLTKSGRK KCSRAAAKSQ ILI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
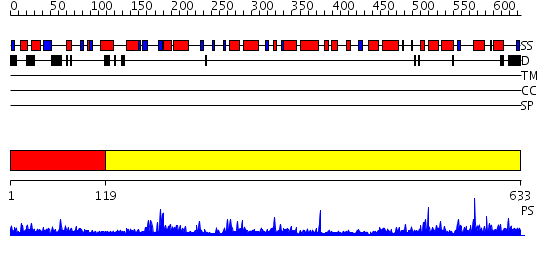
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | 1.259996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [119..633] | 30.0 | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)