













| General Information: |
|
| Name(s) found: |
P2C68_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 393 amino acids |
Gene Ontology: |
|
| Cellular Component: |
protein serine/threonine phosphatase complex
[IEA]
mitochondrion [IEA] |
| Biological Process: |
protein amino acid dephosphorylation
[IEA]
|
| Molecular Function: |
protein serine/threonine phosphatase activity
[IEA][ISS]
catalytic activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSWLARMAL FCLRPMRRYG RMNRDDDDDD DHDGDSSSSG DSLLWSRELE RHSFGDFSIA 60
61 VVQANEVIED HSQVETGNGA VFVGVYDGHG GPEASRYISD HLFSHLMRVS RERSCISEEA 120
121 LRAAFSATEE GFLTLVRRTC GLKPLIAAVG SCCLVGVIWK GTLLIANVGD SRAVLGSMGS 180
181 NNNRSNKIVA EQLTSDHNAA LEEVRQELRS LHPDDSHIVV LKHGVWRIKG IIQVSRSIGD 240
241 AYLKRPEFSL DPSFPRFHLA EELQRPVLSA EPCVYTRVLQ TSDKFVIFAS DGLWEQMTNQ 300
301 QAVEIVNKHP RPGIARRLVR RAITIAAKKR EMNYDDLKKV ERGVRRFFHD DITVVVIFID 360
361 NELLMVEKAT VPELSIKGFS HTVGPSKFSI FLS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
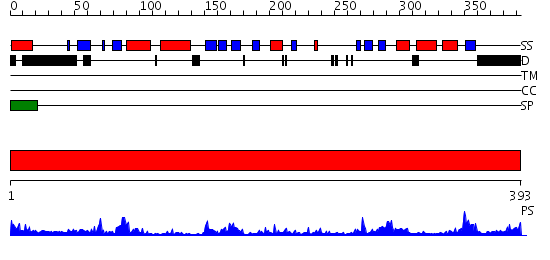
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..393] | 59.522879 | CRYSTAL STRUCTURE OF THE PROTEIN SERINE/THREONINE PHOSPHATASE 2C AT 2 A RESOLUTION |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)