













| General Information: |
|
| Name(s) found: |
mre-11 /
CE37459
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 728 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP germline cell cycle switching, mitotic to meiotic cell cycle [IMP chromosome segregation [IMP DNA metabolic process [IEA double-strand break repair [IEA |
| Molecular Function: |
exonuclease activity
[IEA endonuclease activity [IEA manganese ion binding [IEA hydrolase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCGSDDSFDD FVPDSQEPAS SRTRNQDHLD DDEVPCSQRP DAANDTMVED LDEGDEPAHD 60
61 ESEDIIKILV ATDIHCGYGE NKANIHMDAV NTFEEVLQIA TEQKVDMILL GGDLFHENNP 120
121 SREVQHRVTQ LLRQYCLNGN PIALEFLSDA SVNFNQSVFG HVNYYDQNLN VGLPIFTIHG 180
181 NHDDLSGKGL TALDLLHESG LVNLFGKHSN IQEFIVSPIL LRKGETRLAL YGIGSQRDDR 240
241 LVRAFKNNSI SFLRPNAGAE DWFNLFVLHQ NRPRRAMHRS TGNFLPESLI PQFFDLLIWG 300
301 HEHECKPDPQ YVASSEAVGD GFYILQPGST VATSLTPEEA LQKNAFLIKI KGRKFASKPI 360
361 PLQTVRPMVC DELLLDKIPP GSRPILKTDR PKHTDGRYID EIAIEAKINE MITTAKAKRR 420
421 PRQPELPLIR LKVIYDGDWL NITPANAKRI GLRYENVVAN AVDMVFIKKN NKPKEGKLQT 480
481 ENEKNITEMA DEMGQVSATN LQTIINDYFI NQPLVDQMTV LKPIGIGRAL EQYSAIEEGG 540
541 LATSANRNFD SCLMHQIDVV RKTLKKMPLP PINDPSDLDK FRDLITKDLY DMKKADSERF 600
601 KNPVADVEME EDEDDPQYYM PPQSTSRTNY ASGSEDEVAN SDEEMGSSIS RHSKQPTTRG 660
661 RGRGARGAGA SRETTRGRSN KVVSTRQIDS DGFEIINDSP SPPPASRSTR GKARGKSAPS 720
721 KKRDLSFF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
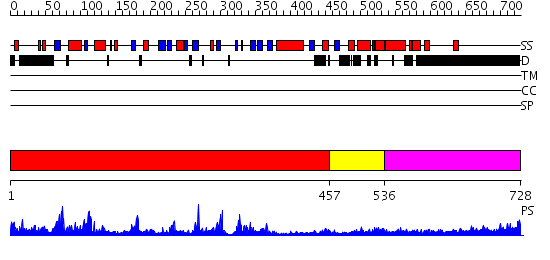
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..456] | 45.39794 | Mre11 | |
| 2 | View Details | [457..535] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [536..728] | 7.098997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)