













| General Information: |
|
| Name(s) found: |
PPOCM_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 508 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA] chloroplast envelope [IDA] plastid [IDA] |
| Biological Process: |
porphyrin biosynthetic process
[TAS]
embryonic development ending in seed dormancy [IMP] |
| Molecular Function: |
oxidoreductase activity
[IEA]
electron carrier activity [IEA] oxygen-dependent protoporphyrinogen oxidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASGAVADHQ IEAVSGKRVA VVGAGVSGLA AAYKLKSRGL NVTVFEADGR VGGKLRSVMQ 60
61 NGLIWDEGAN TMTEAEPEVG SLLDDLGLRE KQQFPISQKK RYIVRNGVPV MLPTNPIELV 120
121 TSSVLSTQSK FQILLEPFLW KKKSSKVSDA SAEESVSEFF QRHFGQEVVD YLIDPFVGGT 180
181 SAADPDSLSM KHSFPDLWNV EKSFGSIIVG AIRTKFAAKG GKSRDTKSSP GTKKGSRGSF 240
241 SFKGGMQILP DTLCKSLSHD EINLDSKVLS LSYNSGSRQE NWSLSCVSHN ETQRQNPHYD 300
301 AVIMTAPLCN VKEMKVMKGG QPFQLNFLPE INYMPLSVLI TTFTKEKVKR PLEGFGVLIP 360
361 SKEQKHGFKT LGTLFSSMMF PDRSPSDVHL YTTFIGGSRN QELAKASTDE LKQVVTSDLQ 420
421 RLLGVEGEPV SVNHYYWRKA FPLYDSSYDS VMEAIDKMEN DLPGFFYAGN HRGGLSVGKS 480
481 IASGCKAADL VISYLESCSN DKKPNDSL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
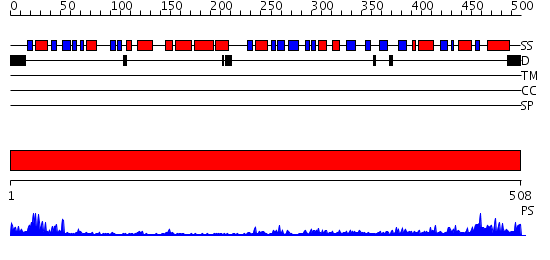
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..508] | 63.045757 | Crystal Structure of Protoporphyrinogen IX Oxidase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)