













| General Information: |
|
| Name(s) found: |
DSP4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 379 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] |
| Biological Process: |
protein amino acid dephosphorylation
[ISS]
starch catabolic process [IMP] starch metabolic process [IMP][TAS] |
| Molecular Function: |
protein tyrosine/serine/threonine phosphatase activity
[ISS]
polysaccharide binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNCLQNLPRC SVSPLLGFGC IQRDHSSSSS SLKMLISPPI KANDPKSRLV LHAVSESKSS 60
61 SEMSGVAKDE EKSDEYSQDM TQAMGAVLTY RHELGMNYNF IRPDLIVGSC LQTPEDVDKL 120
121 RKIGVKTIFC LQQDPDLEYF GVDISSIQAY AKKYSDIQHI RCEIRDFDAF DLRMRLPAVV 180
181 GTLYKAVKRN GGVTYVHCTA GMGRAPAVAL TYMFWVQGYK LMEAHKLLMS KRSCFPKLDA 240
241 IRNATIDILT GLKRKTVTLT LKDKGFSRVE ISGLDIGWGQ RIPLTLDKGT GFWILKRELP 300
301 EGQFEYKYII DGEWTHNEAE PFIGPNKDGH TNNYAKVVDD PTSVDGTTRE RLSSEDPELL 360
361 EEERSKLIQF LETCSEAEV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
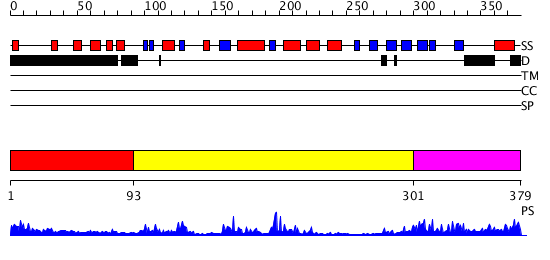
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [93..300] | 29.045757 | Pten tumor suppressor (Phoshphoinositide phosphatase), C-terminal domain; Phoshphoinositide phosphatase Pten (Pten tumor suppressor), N-terminal domain | |
| 3 | View Details | [301..379] | 1.13 | No description for 2qlvB was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.68 |
Source: Reynolds et al. (2008)