













| General Information: |
|
| Name(s) found: |
CTBP_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 636 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
monopolar cell growth
[IMP]
microtubule cytoskeleton organization [IMP] leaf morphogenesis [IMP] |
| Molecular Function: |
protein binding
[IPI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKIRSSATM PHRDQPSPAS PHVVTLNCIE DCALEQDSLA GVAGVEYVPL SRIADGKIES 60
61 ATAVLLHSLA YLPRAAQRRL RPHQLILCLG SADRAVDSTL AADLGLRLVH VDTSRAEEIA 120
121 DTVMALILGL LRRTHLLSRH ALSASGWLGS LQPLCRGMRR CRGMVLGIVG RSVSARYLAS 180
181 RSLAFKMSVL YFDVPEGDEE RIRPSRFPRA ARRMDTLNDL LAASDVISLH CALTNDTVQI 240
241 LNAECLQHIK PGAFLVNTGS CQLLDDCAVK QLLIDGTIAG CALDGAEGPQ WMEAWVKEMP 300
301 NVLILPRSAD YSEEVWMEIR EKAISILHSF FLDGVIPSNT VSDEEVEESE ASEEEEQSPS 360
361 KHEKLAIVES TSRQQGESTL TSTEIVRREA SELKESLSPG QQHVSQNTAV KPEGRRSRSG 420
421 KKAKKRHSQQ KYMQKTDGSS GLNEESTSRR DDIAMSDTEE VLSSSSRCAS PEDSRSRKTP 480
481 LEVMQESSPN QLVMSSKKFI GKSSELLKDG YVVALYAKDL SGLHVSRQRT KNGGWFLDTL 540
541 SNVSKRDPAA QFIIAYRNKD TVGLRSFAAG GKLLQINRRM EFVFASHSFD VWESWSLEGS 600
601 LDECRLVNCR NSSAVLDVRV EILAMVGDDG ITRWID |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
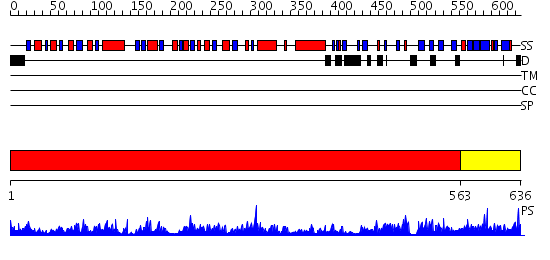
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..562] | 111.0 | Crystal Structure of D-3-Phosphoglycerate dehydrogenase From Mycobacterium tuberculosis | |
| 2 | View Details | [563..636] | 1.085982 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)