













| General Information: |
|
| Name(s) found: |
SUVHA_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 312 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
chromatin modification
[IEA]
|
| Molecular Function: |
zinc ion binding
[IEA]
histone-lysine N-methyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLVGLHSGT IDMEFIGVED HGDEEGKQIA VSVISSGKNA DKTEDPDSLI FTGFGGTDMY 60
61 HGQPCNQKLE RLNIPLEAAF RKKSIVRVVR CMKDEKRTNG NIYIYDGTYM ITNRWEEEGQ 120
121 NGFIVFKFKL VREPDQKPAF GIWKSIQNWR NGLSIRPGLI LEDLSNGAEN LKVCLVNEVD 180
181 KENGPALFRY VTSLIHEVIN NIPSMVDRCA CGRRSCGSKH VFREKLSVSS SLVISAKKSG 240
241 NVARFMNHSC SPNVFWQSIA REQNGLWCLY IGFFAMKHIP PLTELRYDYG KSRGGGKKMC 300
301 LCRTKKCCGS FG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
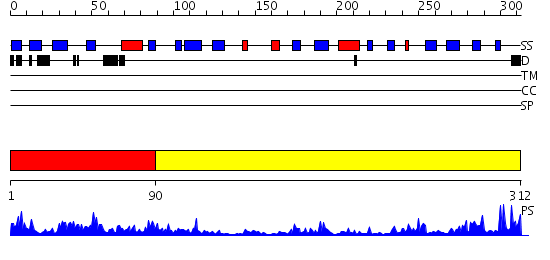
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 1.095991 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [90..312] | 34.522879 | No description for 2igqA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)