













| General Information: |
|
| Name(s) found: |
RGAP7_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 870 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast thylakoid membrane
[IDA]
|
| Biological Process: |
signal transduction
[ISS]
|
| Molecular Function: |
phosphoinositide binding
[ISS]
Rho GTPase activator activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEASLAALER PRGSASNTVF KSGPLFISSK GLGWTSWKKR WFILTRTSLV FFKNDPGTLP 60
61 QKGGEVNLTL GGIDLNNSGS VVVREDKKLL TVLFPDGRDG RAFTLKAETF EDLYEWKTAL 120
121 EQALAQAPNA ALIMGQNGIF RAETNEAIEG REKRPLKSLV VGRPILLALE DIDGSPSFLE 180
181 KALQFIEKYG TKIEGILRQS ADVEEVERRV QEYEQGKTEF TFDEDPHVVG DCIKHVLREL 240
241 PSSPVSASCC TALLEAYRIE SKEARISSLR SAIAETFPEP NRRLLQRILK MMHTISSHSH 300
301 ENRMNPNAVA ACMAPLLLRP LLAGECDLED DFDGGEDNSA QLLAAANAAN NAQAIITVLL 360
361 EDYGSIFDEE NIQRCSISTE SHIGNSGPDD SSDDDNNMKN GYHNADNEVE PVTDDDNDRA 420
421 LSGKMSESSG CTGSDLYEYK GFVADDSDIE SPRDTNGPRC NSNIRTDHLM RNPFVNSTDQ 480
481 QAGEQIGDDP TKYGVNSCLA HVSESYQQSG TGLNVPTHGN TLAAPGLESP SAKSVNKGTP 540
541 SSVHAKRATF WGRGSARKIS TDGSFDSSGE DELAIQRLET TKNELRQRIA KEARGNAILQ 600
601 ASLERRKQAL HERRLSLEQD VSRLQEQLQA ERDLRAALEV GLSMSSGQFS SHGVDSKTRA 660
661 ELEEIALAEA DVARLKQKVA ELHHQLNQQR QTHFGSFSDA RDTHQYLQNH NPQKRFLQQD 720
721 FDSTLAYVNH ERKQRHEENV LGAEWRNSKG AGSFGVGNSR QPSRKQIPES TNTTDSKISE 780
781 ESGKISVDKL SSIDSPSIPS TSRVLDITEY PRLNHPSAAA SAALVELTTR LDFFKERRSQ 840
841 LMEQLQNLDL NYGGSSSQDF IHRPSSPPWN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
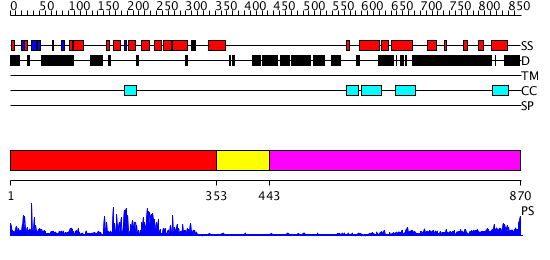
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..352] | 55.30103 | No description for 3cxlA was found. | |
| 2 | View Details | [353..442] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [443..870] | 16.69897 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)