













| General Information: |
|
| Name(s) found: |
HDG2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 721 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS]
trichome morphogenesis [IMP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFEPNMLLAA MNNADSNNHN YNHEDNNNEG FLRDDEFDSP NTKSGSENQE GGSGNDQDPL 60
61 HPNKKKRYHR HTQLQIQEME AFFKECPHPD DKQRKQLSRE LNLEPLQVKF WFQNKRTQMK 120
121 NHHERHENSH LRAENEKLRN DNLRYREALA NASCPNCGGP TAIGEMSFDE HQLRLENARL 180
181 REEIDRISAI AAKYVGKPVS NYPLMSPPPL PPRPLELAMG NIGGEAYGNN PNDLLKSITA 240
241 PTESDKPVII DLSVAAMEEL MRMVQVDEPL WKSLVLDEEE YARTFPRGIG PRPAGYRSEA 300
301 SRESAVVIMN HVNIVEILMD VNQWSTIFAG MVSRAMTLAV LSTGVAGNYN GALQVMSAEF 360
361 QVPSPLVPTR ETYFARYCKQ QGDGSWAVVD ISLDSLQPNP PARCRRRASG CLIQELPNGY 420
421 SKVTWVEHVE VDDRGVHNLY KHMVSTGHAF GAKRWVAILD RQCERLASVM ATNISSGEVG 480
481 VITNQEGRRS MLKLAERMVI SFCAGVSAST AHTWTTLSGT GAEDVRVMTR KSVDDPGRPP 540
541 GIVLSAATSF WIPVPPKRVF DFLRDENSRN EWDILSNGGV VQEMAHIANG RDTGNCVSLL 600
601 RVNSANSSQS NMLILQESCT DPTASFVIYA PVDIVAMNIV LNGGDPDYVA LLPSGFAILP 660
661 DGNANSGAPG GDGGSLLTVA FQILVDSVPT AKLSLGSVAT VNNLIACTVE RIKASMSCET 720
721 A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
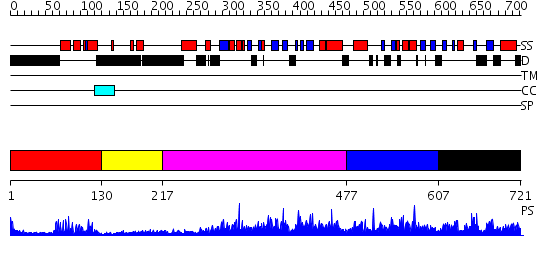
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..129] | 19.045757 | Paired protein | |
| 2 | View Details | [130..216] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [217..476] | 2.71 | No description for 2r55A was found. | |
| 4 | View Details | [477..606] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [607..721] | 3.061986 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)