













| General Information: |
|
| Name(s) found: |
gi|27311575
[NCBI NR]
gi|18420952 [NCBI NR] |
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 714 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] cytosol [IDA] |
| Biological Process: |
translational initiation
[IEA]
|
| Molecular Function: |
nucleic acid binding
[ISS]
translation initiation factor activity [ISS][TAS] protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAVVDIDVL TAQLGINWSD VNLDSIQLPP GNDFGIESDD ESVYQDDQSE FDTGFGNIIV 60
61 VDHLPVVPKE KFEKLEGVVK KIYNQLGVIK ENGLLMPVDP DTKMTLGYCF IEFNTPQEAQ 120
121 NAKEKSHGYK LDKSHIFAVN MFDDFDRLMN VKEEWEPPQE RPYVPGENLQ KWLTDDKARD 180
181 QLVIRFGHDT EVYWNDARQK KPEPVHKRSY WTESYVQWSP IGTYLVTLHK QGAAVWGGAD 240
241 TFTRLMRYQH SMVKLVDFSP GEKYLVTYHS QEPSNPRDAS KVEIKVFDVR TGRMMRDFKG 300
301 SADEFSIGGP GGVAGASWPV FRWAGGKDDK YFAKLSKNTI SVYETETFSL IDKKSMKVDN 360
361 VVDICWSPTD SILSLFVPEQ GGGNQPAKVA LVQIPSKVEL RQKNLFSVSD CKMYWQSSGE 420
421 YLAVKVDRYT KTKKSTYSGF ELFRIKERDI PIEVLELDNK NDKIIAFAWE PKGHRFAVIH 480
481 GDQPRPDVSF YSMKTAQNTG RVSKLATLKA KQANALFWSP TGKYIILAGL KGFNGQLEFF 540
541 NVDELETMAT AEHFMATDIE WDPTGRYVAT SVTTVHEMEN GFTIWSFNGN MVYRILKDHF 600
601 FQLAWRPRPA SFLTAEKEEE IAKNLRNYSK RYEAEDQDVS LLLSEQDREK RRALNEEWQK 660
661 WVMQWKSLHE EEKLVRQNLR DGEVSDVEED EYEAKEVEFE DLIDVTEEIV QELM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
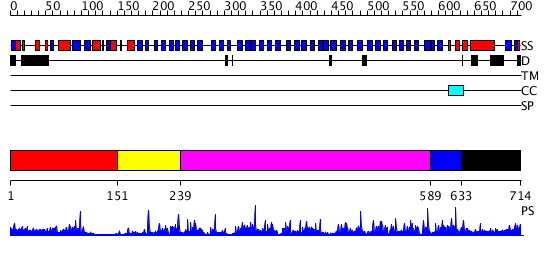
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..150] | 12.69897 | No description for 2nlwA was found. | |
| 2 | View Details | [151..238] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [239..588] | 60.39794 | PAF-AH Holoenzyme: Lis1/Alfa2 | |
| 4 | View Details | [589..632] | 1.708994 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [633..714] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)