













| General Information: |
|
| Name(s) found: |
SPSA3_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1062 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biosynthetic process
[IEA]
sucrose metabolic process [IEA] |
| Molecular Function: |
transferase activity, transferring glycosyl groups
[ISS]
sucrose-phosphate synthase activity [RCA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGNEWINGY LEAILDSQAQ GIEETQQKPQ ASVNLREGDG QYFNPTKYFV EEVVTGVDET 60
61 DLHRTWLKVV ATRNSRERNS RLENMCWRIW HLTRKKKQLE WEDSQRIANR RLEREQGRRD 120
121 ATEDLSEDLS EGEKGDGLGE IVQPETPRRQ LQRNLSNLEI WSDDKKENRL YVVLISLHGL 180
181 VRGENMELGS DSDTGGQVKY VVELARALAR MPGVYRVDLF TRQICSSEVD WSYAEPTEML 240
241 TTAEDCDGDE TGESSGAYII RIPFGPRDKY LNKEILWPFV QEFVDGALAH ILNMSKVLGE 300
301 QIGKGKPVWP YVIHGHYADA GDSAALLSGA LNVPMVLTGH SLGRNKLEQL LKQGRQSKED 360
361 INSTYKIKRR IEAEELSLDA AELVITSTRQ EIDEQWGLYD GFDVKLEKVL RARARRGVNC 420
421 HGRFMPRMAV IPPGMDFTNV EVQEDTPEGD GDLASLVGGT EGSSPKAVPT IWSEVMRFFT 480
481 NPHKPMILAL SRPDPKKNIT TLLKAFGECR PLRELANLTL IMGNRDDIDE LSSGNASVLT 540
541 TVLKLIDKYD LYGSVAYPKH HKQSDVPDIY RLAANTKGVF INPALVEPFG LTLIEAAAHG 600
601 LPMVATKNGG PVDIHRALHN GLLVDPHDQE AIANALLKLV SEKNLWHECR INGWKNIHLF 660
661 SWPEHCRTYL TRIAACRMRH PQWQTDADEV AAQDDEFSLN DSLKDVQDMS LRLSMDGDKP 720
721 SLNGSLEPNS ADPVKQIMSR MRTPEIKSKP ELQGKKQSDN LGSKYPVLRR RERLVVLAVD 780
781 CYDNEGAPDE KAMVPMIQNI IKAVRSDPQM AKNSGFAIST SMPLDELTRF LKSAKIQVSE 840
841 FDTLICSSGS EVYYPGGEEG KLLPDPDYSS HIDYRWGMEG LKNTVWKLMN TTAVGGEARN 900
901 KGSPSLIQED QASSNSHCVA YMIKDRSKVM RVDDLRQKLR LRGLRCHPMY CRNSTRMQIV 960
961 PLLASRSQAL RYLFVRWRLN VANMYVVVGD RGDTDYEELI SGTHKTVIVK GLVTLGSDAL1020
1021 LRSTDLRDDI VPSESPFIGF LKVDSPVKEI TDIFKQLSKA TA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
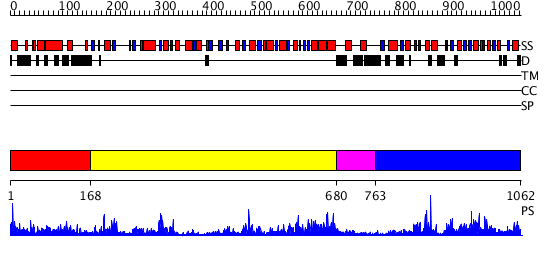
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 1.049982 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [168..679] | 60.0 | No description for 2r60A was found. | |
| 3 | View Details | [680..762] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [763..1062] | 30.69897 | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 at 1.40 A resolution |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)