













| General Information: |
|
| Name(s) found: |
PNP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 922 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] |
| Biological Process: |
chlorophyll biosynthetic process
[IMP]
carotene biosynthetic process [IMP] xanthophyll biosynthetic process [IMP] negative regulation of isopentenyl diphosphate biosynthetic process, mevalonate-independent pathway [IMP] |
| Molecular Function: |
nucleic acid binding
[ISS]
3'-5'-exoribonuclease activity [IEA][ISS] RNA binding [IEA][ISS] polyribonucleotide nucleotidyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLTSPSNALH SSTPQFWPLR RSKLCRSRNF PRFHSGERSS GGGGKLCSLS LLSGSGAGKF 60
61 SVRALVRPDD TDDADSVGDG SLAFPNHVSV KIPFGNREIL VETGLMGRQA SSAVTVTDGE 120
121 TIVYTSVCLA DVPSEPSDFL PLYVHYQERF SAVGRTSGGF FKREGRTKDH EVLICRLIDR 180
181 PLRPTMPKGF YNETQILSWV LSYDGLHAPD ALAVTSAGIA VALSEVPNAK AIAGVRVGLI 240
241 GGEFIVNPTV KEMEESQLDL FLAGTDTAIL TIEGYSNFLP EEMLLQAVKV GQDAVQATCI 300
301 AIEVLAKKYG KPKMLDAIRL PPPELYKHVK ELAGEELTKA LQIKSKISRR KAISSLEEKV 360
361 LTILTEKGYV IDEVAFGTIE AQPDLLEDED EDEEVVPEGE VDQGDVHIRP IPRKPIPLLF 420
421 SEVDVKLVFK EVSSKLLRRR IVEGGKRSDG RTLDEIRPIN SRCGLLPRAH GSTLFTRGET 480
481 QALAVVTLGD KQMAQRIDNL EGSDEYKRFY LQYTFPPSSV GEVGRIGAPS RREIGHGTLA 540
541 ERALETILPS DDDFPYTIRV ESTVIESNGS SSMASVCGGC LALQDAGVPV KCSVAGIAMG 600
601 MVWDTEEFGG DGSPLILSDI TGAEDASGDM DFKVAGNEDG VTAFQMDIKV GGITLEIMEK 660
661 ALIQAKAGRR HILAEMAKCS PPPTLSLSKY APLILIMKVH PSKVYSLIGS GGKKVKSIIE 720
721 ESGVEAIDMQ DDGTVKIMAI DVASLERAKA IISGLTMVPS VGDIYRNCEI KSMAPYGAFV 780
781 EIAPGREGLC HISELSAEWL AKPEDAYKVG DRIDVKLIEV NEKGQLRLSV RALLPESETD 840
841 KDSQKQQPAG DSTKDKSSQR KYVNTSSKDR AAAGASKVSS GDELVLKKKD VRRATGGSSD 900
901 KTMNSNSSTN EESLVNGEAT IS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
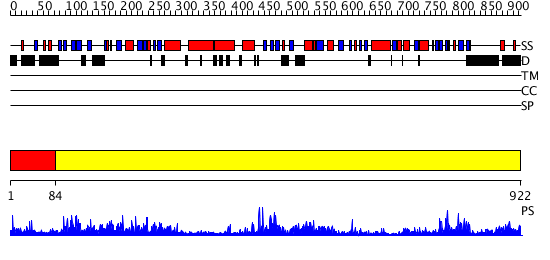
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..83] | 7.0 | Solution structure of the alpha-helical domain from mouse hypothetical PNPase | |
| 2 | View Details | [84..922] | 161.0 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.52 |
Source: Reynolds et al. (2008)