













| General Information: |
|
| Name(s) found: |
TPPD_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 369 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
metabolic process
[IEA]
trehalose biosynthetic process [IEA][ISS] |
| Molecular Function: |
catalytic activity
[IEA]
trehalose-phosphatase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTNHNALISD AKGSIGVAVR VPNQSLFSPG GGRYISIPRK KLVQKLEADP SQTRIHTWIE 60
61 AMRASSPTRT RPGNISPLPE SDEEDEYSSW MAQHPSALTM FEEIAEASKG KQIVMFLDYD 120
121 GTLSPIVENP DRAYMSEEMR EAVKGVARYF PTAIVTGRCR DKVRRFVKLP GLYYAGSHGM 180
181 DIKGPSKRNK HNKNNKGVLF QAANEFLPMI DKVSKCLVEK MRDIEGANVE NNKFCVSVHY 240
241 RCVDQKDWGL VAEHVTSILS EYPKLRLTQG RKVLEIRPTI KWDKGKALEF LLESLGFANS 300
301 NDVLPIYIGD DRTDEDAFKV LRNKGQGFGI LVSKIPKETS ATYSLQEPSE VGEFLQRLVE 360
361 WKQMSLRGR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
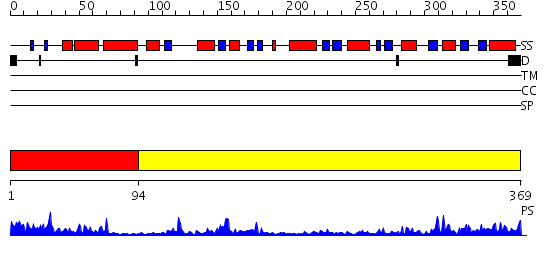
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | 16.221849 | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | |
| 2 | View Details | [94..369] | 31.221849 | Crystal structure of NYSGRC target T1436: A Hypothetical protein yidA. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)