













| General Information: |
|
| Name(s) found: |
TYRA1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 640 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
tyrosine biosynthetic process
[IEA][ISS]
metabolic process [IEA] |
| Molecular Function: |
catalytic activity
[IEA]
binding [IEA] prephenate dehydrogenase (NADP+) activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAETLITKPP LSLSFTSLSS MLPSLSLSTA NRHLSVTDTI PLPNSNSNAT PPLRIAIIGF 60
61 GNYGQFLAET LISQGHILFA HSRSDHSSAA RRLGVSYFTD LHDLCERHPD VVLLCTSILS 120
121 IENILKTLPF QRLRRNTLFV DVLSVKEFAK TLLLQYLPED FDILCTHPMF GPQSVSSNHG 180
181 WRGLRFVYDK VRIGEERLRV SRCESFLEIF VREGCEMVEM SVTDHDKFAA ESQFITHTLG 240
241 RLLGMLKLIS TPINTKGYEA LLDLAENICG DSFDLYYGLF VYNNNSLEVL ERIDLAFEAL 300
301 RKELFSRLHG VVRKQSFEGE AKKVHVFPNC GENDASLDMM RSEDVVVKYE YNSQVSGSVN 360
361 DGSRLKIGIV GFGNFGQFLG KTMVKQGHTV LAYSRSDYTD EAAKLGVSYF SDLDDLFEEH 420
421 PEVIILCTSI LSTEKVLESL PFQRLKRSTL FVDVLSVKEF PRNLFLQTLP QDFDILCTHP 480
481 MFGPESGKNG WNNLAFVFDK VRIGMDDRRK SRCNSFLDIF AREGCRMVEM SCAEHDWHAA 540
541 GSQFITHTVG RLLEKLSLES TPIDTKGYET LLKLVENTAG DSFDLYYGLF LYNPNAMEQL 600
601 ERFHVAFESL KTQLFGRLHS QHSHELAKSS SPKTTKLLTS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
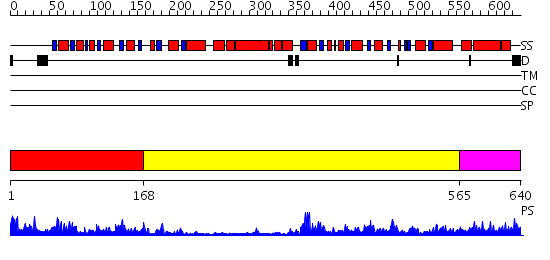
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 42.30103 | Phosphoglycerate dehydrogenase; Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain | |
| 2 | View Details | [168..564] | 16.221849 | K185N mutated S-adenosylhomocysteine hydrolase | |
| 3 | View Details | [565..640] | 41.045757 | Crystal Structure of Prephenate Dehydrogenase from Aquifex aeolicus |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||
| 3 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|