













| General Information: |
|
| Name(s) found: |
SUVR1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 630 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA]
|
| Biological Process: |
chromatin modification
[IEA]
|
| Molecular Function: |
zinc ion binding
[IEA]
histone-lysine N-methyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDEDEFPLKR RLRSRRGRAS SSSSSSSSYN NEDLKTQPEE EDEDDGVTEL PPLKRYVRRN 60
61 GERGLAMTVY NNASPSSSSR LSMEPEEVPP MVLLPAHPME TKVSEASALV ILNDEPNIDH 120
121 KPVISDTGNC SAPMLEMGKS NIHVQEWDWE TKDILNDTTA MDVSPSSAIG ESSEHKVAAA 180
181 SVELASSTSG EAKICLSFAP ATGETTNLHL PSMEDLRRAM EEKCLKSYKI VHPEFSVLGF 240
241 MKDMCSCYID LAKNSTSQLL ETETVCDMSK AGDESGAVGI SMPLVVVPEC EISGDGWKAI 300
301 SNMKDITAGE ENVEIPWVNE INEKVPSRFR YMPHSFVFQD APVIFSLSSF SDEQSCSTSC 360
361 IEDCLASEMS CNCAIGVDNG FAYTLDGLLK EEFLEARISE ARDQRKQVLR FCEECPLERA 420
421 KKVEILEPCK GHLKRGAIKE CWFKCGCTKR CGNRVVQRGM HNKLQVFFTP NGKGWGLRTL 480
481 EKLPKGAFIC EYIGEILTIP ELYQRSFEDK PTLPVILDAH WGSEERLEGD KALCLDGMFY 540
541 GNISRFLNHR CLDANLIEIP VQVETPDQHY YHLAFFTTRD IEAMEELAWD YGIDFNDNDS 600
601 LMKPFDCLCG SRFCRNKKRS TKTMQILNKA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
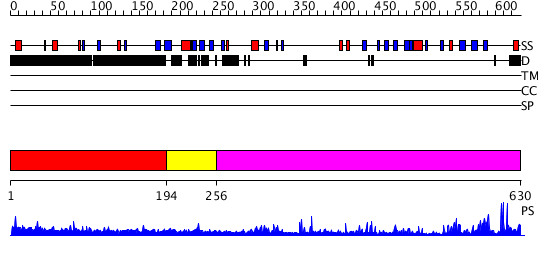
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..193] | 2.069988 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [194..255] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [256..630] | 42.045757 | No description for 2o8jA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)