













| General Information: |
|
| Name(s) found: |
gi|220956804
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | synthetic construct |
| Length: | 713 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLQSSNPNFR SLDLSLGTPT PTTPTSSEIP AAPASATASL SGSTLQFGPE EPEEGSAAGG 60
61 SQSLTPLDAR ELLTRPTRAY ASVSQPQSPR HRGSGCGSVS GSRIITHGLT GRSTSISSQL 120
121 EKTKKLLKMT EGEDKPLTIL HYNDVYNIES MAETEPVGGA ARFATAIKSF AHLNPLVLFS 180
181 GDAFSPSMLS TFTQGEQMIP VLNTVGTHCA VFGNHDFDHG LDVLVKLIKQ TEFPWLMSNV 240
241 VDNETGRPLG GGKISHFILH NQISIGLIGL VEREWLETLP TIDPNEVTYI DYVEAGNKLA 300
301 RELRNEGCDL IIALTHMRTP NDINLAEKCN GIDIILGGHD HVREVTEING KMIVKSGTDF 360
361 QQFSVITIER DAANREHFTT DVKCFDVTAK IPEDPELKQE LSKYAKFIES KLSDVMGVFS 420
421 VELDGRFSRV RTQETNLGNW VCDVVLAAVG ADVVILNGGT FRSDRVHPVG AFTMGDLVNV 480
481 IPMRDPLILL EVKGKILWQA LENGVSAYPK LEGRFPQVSG ISFAFDPQAE PGKRIDPQLI 540
541 QVGDEYLNLE QSYKLCVKSY IFMGCDGYTM FKDATVLMDD DACPELGITL QNHFKAINSR 600
601 KCGQNTKHRQ SLVTLSRRHS LVQCLDSMDL DGPSPIRKLS VGHHNKSMDL THGNSQKMLR 660
661 RASLDDLEQS TCDLAPQLEH RIVMIQNEEH HRQLLFKKET HIMNSTITEA EDE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
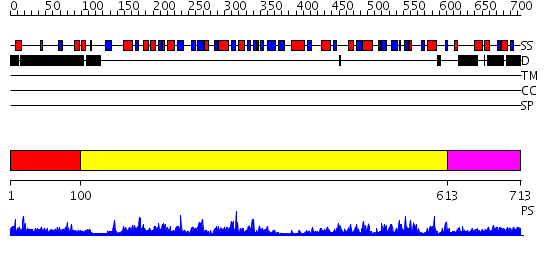
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | 3.077994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [100..612] | 102.0 | 5'-NUCLEOTIDASE FROM E. COLI | |
| 3 | View Details | [613..713] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)