













| General Information: |
|
| Name(s) found: |
gi|190408739
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 753 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVEFSLKKAR NNWKHVKKSA SSPAKQKTPP SPAKPKQKTK KNPYSDLKDP ATSYTLPTIN 60
61 ARERSRVATS MQRRLSIHNT NYAPPTLDYS MPLPDMPNMI VPNDNVDSSH NNSSFTTENE 120
121 SVSSKGPSNS LNLSTADLSL NDSSYNKVPA RSAMRNTVNP SGSNDPFNNS TSLRKMLANP 180
181 HFNAKDFVHD KLGNASAITI DKFTSNLTDL SIQVQEEVKL NINKSYNEIM TVNNDLNVAM 240
241 LELKRVRANI NDLNEVLDQC TKIAEKRLQL QDQIDQERQG NFNNVESHSN SPALLPPLKA 300
301 GQNGNLMRRD RSSVLILEKF WDTELDQLFK NVEGAQKFIN STKGRHILMN SANWMELNTT 360
361 TGKPLQMVQI FILNDLVLIA DKSRDKQNDF IVSQCYPLKD VTVTQEEFST KRLLFKFSNS 420
421 NSSLYECRDA DECSRLLDVI RKAKDDLCDI FHVEEENSKR IRESFRYLQS TQQTPGRENN 480
481 RSPNKNKRRS MGGSITPGRN VTGAMDQYLL QNLTLSMHSR PRSRDMSSTA QRLKFLDEGV 540
541 EEIDIELARL RFESAVETLL DIESQLEDLS ERISDEELML LNLISLKIEQ RREAISSKLS 600
601 QSILSSNEIV HLKSGTENMI KLGLPEQALD LFLQNRSNFI QDLILQIGSV DNPTNYLTQL 660
661 AVIRFQTIKK TVEDFQDIFK ELGAKISSIL VDWCSDEVDN HFKLIDKQLL NDEMLSPGSI 720
721 KSSRKQIDGL KAVGLDFVYK LDEFIKKNSD KIR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
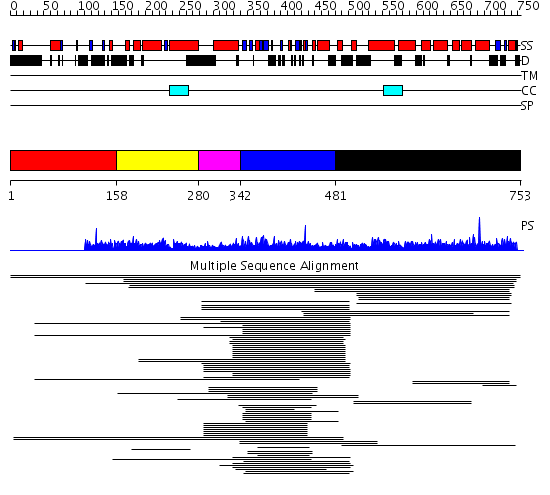
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [158..279] | 1.015997 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [280..341] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [342..480] | 3.09691 | Son of sevenless-1 (sos-1) | |
| 5 | View Details | [481..753] | 4.017997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)