













| General Information: |
|
| Name(s) found: |
gi|194449488
[NCBI NR]
gi|194407792 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Salmonella enterica subsp. enterica serovar Heidelberg str. SL476 |
| Length: | 638 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSDIKIKVQ SFGRFLSNMV MPNIGAFIAW GIITALFIPT GWLPNETLAK LVGPMITYLL 60
61 PLLIGYTGGR LVGGERGGVV GAITTMGVIV GADMPMFLGS MIAGPLGGYC IKKFDSWVDG 120
121 KIKSGFEMLV NNFSAGIIGM ILAILAFLGI GPAVEVLSKI LAAGVNFMVA HDMLPLASIF 180
181 VEPAKILFLN NAINHGIFSP LGIQQSHEMG KSIFFLIEAN PGPGMGVLLA YMFFGRGSAK 240
241 QSAGGAAIIH FLGGIHEIYF PYVLMNPRLI LAVILGGMTG VFTLTILNGG LVSPASPGSI 300
301 LAVLAMTPKG AYFANIAAIV AAMAVSFVVS AILLKTSKVK EEDDIEAATR RMHDMKAESK 360
361 GASPLAAGDV TNDLSHVRKI IVACDAGMGS SAMGAGVLRK KVQDAGLSQI SVTNSAINNL 420
421 PPDVDLVITH RDLTERAMRQ VPQAQHISLT NFLDSGLYTS LTERLVAAQR HNTNEEKVRD 480
481 HLKDSFEEGD NNLFKLGAEN IFLGRKAANK EEAIRFAGEQ LVKGGYVEPE YVEAMLDREK 540
541 LTPTYLGESI AVPHGTVEAK DRVLKTGVVF CQYPEGVRFG EEEDDIARLV IGIAARNNEH 600
601 IQVITSLTNA LDDESVIERL AHTTSVDEVL ELLAGKKA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
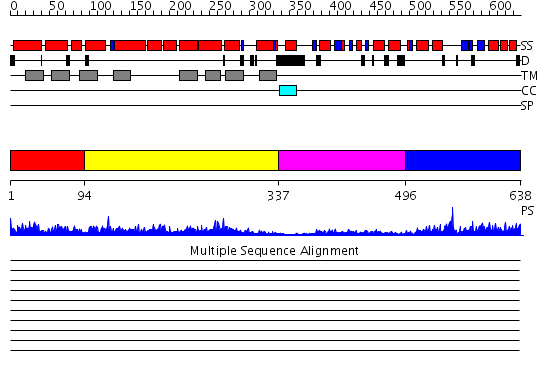
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | 3.621996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [94..336] | 3.637996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [337..495] | 16.30103 | STRUCTURE OF IIB DOMAIN OF THE MANNITOL-SPECIFIC PERMEASE ENZYME II | |
| 4 | View Details | [496..638] | 37.0 | Phosphotransferase IIa-mannitol |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | |||||||||||||||||||||||||||||||||||||
| 4 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|