













| General Information: |
|
| Name(s) found: |
gi|190407993
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 1065 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDYLKPAQKG RRRGLSINSL SETQQSAMNS SLDHLQNDLN RINLQWNRIL SDNTNPLELA 60
61 LAFLDDTSVG LGHRYEEFNQ LKSQIGSHLQ DVVNEHSQVF NTNVASYGKA VSSIMQAQEQ 120
121 TLNLKNCLKE ANEKITTDKG SLQELNDNNL KYTKMIDVLV NIEELLQIPE KIEENIRKEN 180
181 FHQVQILLER GFILMNNKSL KTVEILKPIN QQLELQEHLL FNNLIEEIHD IMYSKSNKTN 240
241 FTRVTNNDIF KIISISHNGF TSLENYLYNI VNIDIMEHSK TINKNLEQFI HDQSLNKGNI 300
301 MLQENAATQA PLAPSRNQEN EGFNRIGFLL KTINNINKLP VAFNIITERA KEEIHNIIVK 360
361 STESIRSKHP SLLKMATSLK NDNHFGLPVQ DILSIILREC FWEIFLKLLY AIQCHRAIFE 420
421 MSNILQPTSS AKPAFKFNKI WGKLLDEIEL LLVRYINDPE LISSNNGSIK PINGATNNAP 480
481 TLPKRKNPKI FSLEYNIEDN SSVKDQAFEL KALLKDIFPG FSVSSNMDLD SIYVKDESFE 540
541 QDEPLVPPSV FNMKVILDPF LLFTQSTSTI VPSVLTQNTI SSLTFFDDYM NKSFLPKIQM 600
601 TMDYLFTVEV ESNNPYALEL SDENHNIFKT ALDFQRLFYN LLNVFNTANT FREKISYCIL 660
661 DLLNHFYNYY LGLFNSLIGT SDRHLTRKII TAWLQNGILM DQEQKILNGD ETLFHEESIE 720
721 LFKEIPHFYQ AGKGLSKSDL FNNLTLDTIL QFSASVLWIL NWLPGLKKAI NIDEVSQEPM 780
781 LDADRLRSSW TFSESMDLNY SNPSSSPNSL GNLKILLDDK ASKKFDETID GFKTLKFKLI 840
841 TILRFNIRAL CIYDIGSFFQ NTKIWNMDVG SIELDQNIAS LISELRRTES KLKQQLPEKE 900
901 KNSIFIGLDI VNNYALIKGA KSIKVLNHNG IKKMLRNVNV LQHAYRNLSS EPSKINMNVT 960
961 MNFYSLCGSS EAELFEYIKD NELPHCSVED LKTILRLQFS EEMHRQLKRQ STSSTKGSIK1020
1021 PSNKRYTEAL EKLSNLEKEQ SKEGARTKIG KLKSKLNAVH TANEK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
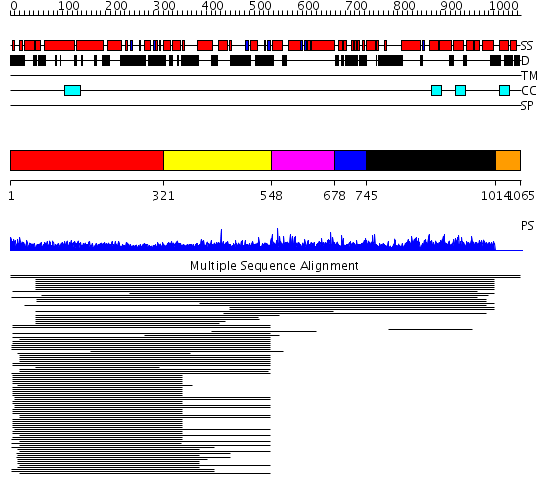
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..320] | 12.69897 | Heavy meromyosin subfragment | |
| 2 | View Details | [321..547] | 2.317989 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [548..677] | 1.015981 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [678..744] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [745..1013] | 1.013948 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1014..1065] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)