













| General Information: |
|
| Name(s) found: |
gi|170023427
[NCBI NR]
gi|169749961 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Yersinia pseudotuberculosis YPIII |
| Length: | 545 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKPTLLLHTS YDMQSSSEHI SDVLIIGSGA AGLSLALRLA PHCKVTVLSK GPLNEGATFY 60
61 AQGGIAAVFD ETDSISSHVD DTLIAGAGLC DKEAVEFIAS NARSCVQWLI DQGVLFDTET 120
121 SANGEERYHL TREGGHSHRR ILHTADATGK EVETTLVGKA SNHPNISVKE RCNAVDLITS 180
181 NKIGLAGTKR VVGAYVWNRE LEKVETFRAK AVVLATGGAA KVYQYTTNPD ISSGDGIAMA 240
241 WRAGCRVANL EFNQFHPTCL FHPQARNFLL TEALRGEGAY LKRPDGSRFM LDFDPRGELA 300
301 PRDIVARAID HEMKRLGADC MYLDISHKPT DFVMQHFPMI YEKLLSLGID LTKEAIPIVP 360
361 AAHYTCGGVM VDQHGRTDLD GLYAIGEVSY TGLHGANRMA SNSLLECLVY GWSAAEDILT 420
421 RLPTAKLAKH LPDWDESRVD NSDERVVIQH NWHELRLFMW DYVGIVRTTK RLERALRRIN 480
481 TLQQEIDEYY ANFRISNNLL ELRNLVQVAE LIVRCAMERK ESRGLHYTLD YPDQIENPQP 540
541 TILHP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
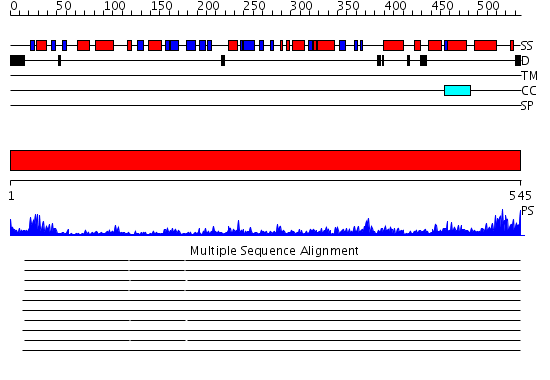
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..545] | 131.0 | L-aspartate oxidase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)