













| General Information: |
|
| Name(s) found: |
gi|207345134
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae AWRI1631 |
| Length: | 1146 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDYFSSRPS QTLTPMGNKP SGGGGGDDAS SIHSKSSQYL MDILPDSMTL NESVSSIVAN 60
61 NQAKEFILPE TDERSPYFIN VPIPKAQPTS TTETKKPLAG DEAIDGQFVK EYPTDILVDR 120
121 FYKWKKILKG LVIYLREVAY AQEQFARINY QLKGSVKFPF LTDIDETTNT ITDPFTTAPR 180
181 GPKKAQPAQK KVGLTDSEQF QMQMQQEQQE NAVQAPTDES KMSLAPHEYK PVQTTESDNT 240
241 SAASGFVKFG SGSIQDIQVI LKKYHLSLAN QQFKISKEIT STVIPKLEEL RKDLRYKITE 300
301 IKDLHGDFKT NIGAHIQLTS QLLKKYIAAV KFMNAHGIGN DRASPTNKKP HKLDPKHDPY 360
361 LLKLQLDLQL KRQVAEETYL QEAFINLQSS GLQLEKIIYT KIQHALLRYS ALIDSEARLM 420
421 IKNMCQELQH GIISKPPAFE WDNFVTQHPS CLLNWKSNDP IPPPRKVSDV IYPHMKSPLA 480
481 KCIKAGYFLK KSELLPTYHQ GYFVLTSNYI HEFQSSDFYN LSSSTPNSTK SSAYSSSVSI 540
541 ADTYANANNA KANNHHRQAS DVHNSSTTTG GTAGANGIRG IRKKSYLAPI MSIPLNDCTL 600
601 KDASSTKFVL VGKPTLNENA DVRKSSSSTY LSGSSQASLP KYGHETAKIF SKAPFHKFLK 660
661 GSKPKNKNTK SSELDQFYAA AQKESNNYVT WTFKIVSPEP SEEELKHFKR WVQDLKNLTS 720
721 FNDTKDRIKF IEDRVMKSHR FKAGHMSRNS VNIGSHTPCL TDSTFTLQDG TTTSVNLKGR 780
781 AEKPQYIHIQ NNSLADFDGN GFRSKVNTPA IDDYGNLITV ERRPAQSPHQ YSDYMATSGN 840
841 TTPSYSSGSR PQSMYNGYNP AVSITSNGMM LQQSTANNNT NPTTNLRHQR NISQTSSLPG 900
901 FSYTSLSLPV NSPGSSNSES SSGGYFAIPL HGNNNNNNYT QRNSEGSSPC YNDDQIRQQQ 960
961 QPLQMQPLSR TSSSSVNVTA MRSTSAGNSI TANAPVVPKV MVNNQNVKTV AADQSATAPS1020
1021 SPTMNSSVTT INRESPYQTL KKTNSTGNVP CLTAEKTHAH PAFYKRGNNS AQNLTTSSST1080
1081 ASRVHPIRKH KKNVSFSSLN SLMFSKKGAN HGGNLMTNQF MSGGIQEDDG DSTNNDTIKL1140
1141 NQSIYS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
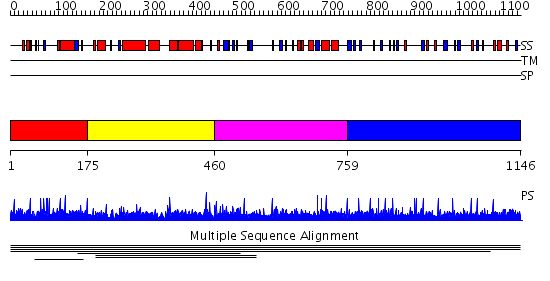
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..174] | 1.005997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [175..459] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [460..758] | 5.886057 | PH domain No confident structure predictions are available. | |
| 4 | View Details | [759..1146] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)