













| General Information: |
|
| Name(s) found: |
gi|207346648
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae AWRI1631 |
| Length: | 971 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDRFQIGDEQ LLRFYQLKTI NPTHSWAQDS SKLNNEEATS NELGVETSFD ILKDFKYGNQ 60
61 ISIDKESRAY LNDESLSYIR DPLNGQEMSK ELQHLPNDSM RLNYLVNSKQ FNVKAFLRDM 120
121 HKQDSFNDLN NSLDRLDSDI QDQSIHLKQL VGKNFTKYVK IKNKLDQIYK EFDEKTNEKN 180
181 QCDSPKENQI NVESLNKKVD EVIRTTTFKL KPLMDNYQKI LNYQATKKFI ELNKFYFNLP 240
241 KSLKRCLTNN DFNEFIIEYS KGLTLRRRFN QSSDASQSLV IKRIWTQIEN LLVTYKDLIW 300
301 NSLINSNFNI DQPQETILSL FSKLLNLENF INNNQRESES GNKNTTSSSN ENPILRWMSI 360
361 KMNGFQNELN ELSGHMISKI IHSQRLILQN NTNQDKSQGC VELSYYLKIN QLFQIISDTG 420
421 KDSEGLKSTV EPNKVNTISG TSYLNLNCQP SSQGLTDSPT IIEMWLLILK YINDLWKICD 480
481 QFIEFWEHIE KFLDGTYQNS IINEKRKENI LIGDSNIIES YQKSLILKEE QINEVRLKGE 540
541 EFITSVSQNL ISFFTSSQSS LPSSLKDSTG DITRSNKDSG SPLDYGFIPP NCNGLSCLRY 600
601 LPKIVEPILK FSTELAQLNI TTNGITICRN TLSTIINRCV GAISSTKLRD ISNFYQLENW 660
661 QVYETVTFSS KSQDSSKNLT FEYGVTQFPE IVTSFQEVSI KTTRDLLFAY EKLPIINGIS 720
721 VVSYPSKQLL TGIEIQQIIS MEAVLEAILK NAAKDKDNPR NSHTILTLTN LQYFRECAFP 780
781 NILQYFDDAF EWNLASKNLE LFSLLSKMES SIFGNYLSDL KINLRDTLEE KFHEINWPMY 840
841 TSNSFRVGDY IIEALMILIV VHSECFRIGP QLIHKILIET QIFIARYLFE AFKPYVGNLS 900
901 NDGSLQIIVD LEFFQKVMGP LLEKDTEATL RACLQNCFQN DTNRLQKCIN EINPIVSANL 960
961 KRTAIQFAAF S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
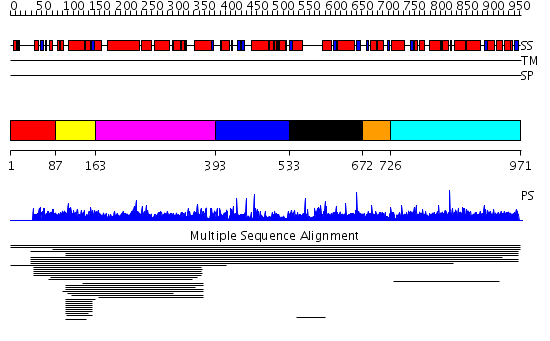
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [87..162] | 5.020992 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [163..392] | 1.016971 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [393..532] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [533..671] | 1.006992 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [672..725] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [726..971] | 1.006942 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)