













| General Information: |
|
| Name(s) found: |
gi|190404822
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 1358 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPNDNKTPNR SSTPKFTKKP VTPNDKIPER EEKSNEVKTP KIPLFTFAKS KNYSRPSTAI 60
61 HTSPHQPSDV KPTSHKQLQQ PKSSPLKKNN YNSFPHSNLE KISNSKLLSL LRSKTSAGRI 120
121 ESNNPSHDAS RSLASFEQTA FSRHAQQQTS TFNSKPVRTI VPISTSQTNN SFLSGVKSLL 180
181 SEEKIRDYSK EILGINLANE QPVLEKPLKK GSADIGASVI SLTKDKSIRK DTVEEKKEEK 240
241 LNIGKNFAHS DSLSVPKVSA GDSGISPEES KARSPGIAKP NAIQTEVYGI NEESTNERLE 300
301 INQEKPVKLD ENSANSTVAS ALDTNGTSAT TETLTSKKIV PSPKKVAIDQ DKITLHDEKT 360
361 LAPSKHQPIT SEQKMKEDAD LKRMEILKSP HLSKSPADRP QGRRNSRNFS TRDEETTKLA 420
421 FLVEYEGQEN NYNSTSRSTE KKNDMNTSAK NKNGENKKIG KRPPEIMSTE AHVNKVTEET 480
481 TKQIQSVRID GRKVLQKVQG ESHIDSRNNT LNVTPSKRPQ LGEIPNPMKK HKPNEGRTPN 540
541 ISNGTINIQK KLEPKEIVRD ILHTKESSNE AKKTIQNPLN KSQNTALPST HKVTQKKDIK 600
601 IGTNDLFQVE SAPKISSEID RENVKSKDEP VSKAVESKSL LNLFSNVLKA PFIKSESKPF 660
661 SSDALSKEKA NFLETIASTE KPENKTDKVS LSQPVSASKH EYSDNFPVSL SQPSKKSFAN 720
721 HTEDEQIEKK KICRGRMNTI ITHPGKMELV YVSDSDDSSS DNDSLTDLES LSSGESNEIK 780
781 VTNDLDTSAE KDQIQAGKWF DPVLDWRKSD RELTKNILWR IADKTTYDKE TITDLIEQGI 840
841 PKHSYLSGNP LTSVTNDICS VENYETSSAF FYQQVHKKDR LQYLPLYAVS TFENTNNTEK 900
901 NDVTNKNINI GKHSQEQNSS SAKPSQIPTV SSPLGFEETK LSTTPTKSNR RVSHSDTNSS 960
961 KPKNTKENLS KSSWRQEWLA NLKLISVSLV DEFPSELSDS DRQIINEKMQ LLKDIFANNL1020
1021 KSAISNNFRE SDIIILKGEI EDYPMSSEIK IYYNELQNKP DAKKARFWSF MKTQRFVSNM1080
1081 GFDIQKSCEP VSISTSVKPH VVEPEHMADA KIMPKDILQI TKKPLMVKNV KPSSPPDVKS1140
1141 LVQLSTMETK TLPEKKQFDS IFNSNKAKII PGNGKHASEN ISLSFSRPAS YGYFSVGKRV1200
1201 PIVEDRRVKQ LDDITDSNTT EILTSVDVLG THSQTGTQQS NMYTSTQKTE LEIDNKDSVT1260
1261 ECSKDMKEDG LSFVDIVLSK AASALDEKEK QLAVANEIIR SLSDEVMRNE IRITSLQGDL1320
1321 TFTKKCLENA RSQISEKDAK INKLMEKDFQ VNKEIKPY |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
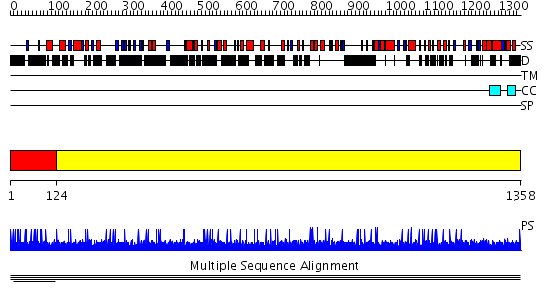
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..123] | 1.002954 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [124..1358] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [277..390] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [391..444] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [445..659] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [660..784] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [785..924] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [925..1081] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1082..1201] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1202..1358] | 78.69897 | Dimerization motif of sir4 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)