













| General Information: |
|
| Name(s) found: |
gi|168272888
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | synthetic construct |
| Length: | 772 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSLKKYLTE GLLQFTILLS LIGVRVDVDT YLTSQLPPLR EIILGPSSAY TQTQFHNLRN 60
61 TLDGYGIHPK SIDLDNYFTA RRLLSQVRAL DRFQVPTTEV NAWLVHRDPE GSVSGSQPNS 120
121 GLALESSSGL QDVTGPDNGV RESETEQGFG EDLEDLGAVA PPVSGDLTKE DIDLIDILWR 180
181 QDIDLGAGRE VFDYSHRQKE QDVEKELRDG GEQDTWAGEG AEALARNLLV DGETGESFPA 240
241 QVPSGEDQTA LSLEECLRLL EATCPFGENA EFPADISSIT EAVPSESEPP ALQNNLLSPL 300
301 LTGTESPFDL EQQWQDLMSI MEMQAMEVNT SASEILYSAP PGDPLSTNYS LAPNTPINQN 360
361 VSLHQASLGG CSQDFLLFSP EVESLPVASS STLLPLAPSN STSLNSTFGS TNLTGLFFPP 420
421 QLNGTANDTA GPELPDPLGG LLDEAMLDEI SLMDLAIEEG FNPVQASQLE EEFDSDSGLS 480
481 LDSSHSPSSL SSSEGSSSSS SSSSSSSSSA SSSASSSFSE EGAVGYSSDS ETLDLEEAEG 540
541 AVGYQPEYSK FCRMSYQDPA QLSCLPYLEH VGHNHTYNMA PSALDSADLP PPSALKKGSK 600
601 EKQADFLDKQ MSRDEHRARA MKIPFTNDKI INLPVEEFNE LLSKYQLSEA QLSLIRDIRR 660
661 RGKNKMAAQN CRKRKLDTIL NLERDVEDLQ RDKARLLREK VEFLRSLRQM KQKVQSLYQE 720
721 VFGRLRDENG RPYSPSQYAL QYAGDGSVLL IPRTMADQQA RRQERKPKDR RK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
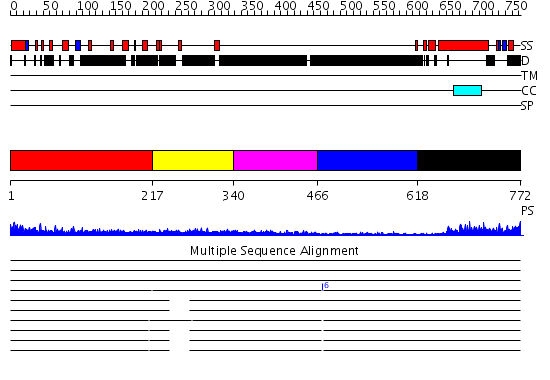
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..216] | 4.234995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [217..339] | 3.237991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [340..465] | 1.218992 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [466..617] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [618..772] | 10.522879 | No description for 2i1jA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.39 |
Source: Reynolds et al. (2008)