













| General Information: |
|
| Name(s) found: |
GWD1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1399 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] mitochondrion [IDA] chloroplast envelope [IDA] |
| Biological Process: |
response to symbiotic fungus
[IEP]
cold acclimation [IMP] starch catabolic process [IMP] |
| Molecular Function: |
alpha-glucan, water dikinase activity
[IMP][TAS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNSVVHNLL NRGLIRPLNF EHQNKLNSSV YQTSTANPAL GKIGRSKLYG KGLKQAGRSL 60
61 VTETGGRPLS FVPRAVLAMD PQAAEKFSLD GNIDLLVEVT STTVREVNIQ IAYTSDTLFL 120
121 HWGAILDNKE NWVLPSRSPD RTQNFKNSAL RTPFVKSGGN SHLKLEIDDP AIHAIEFLIF 180
181 DESRNKWYKN NGQNFHINLP TERNVKQNVS VPEDLVQIQA YLRWERKGKQ MYNPEKEKEE 240
241 YEAARTELRE EMMRGASVED LRAKLLKKDN SNESPKSNGT SSSGREEKKK VSKQPERKKN 300
301 YNTDKIQRKG RDLTKLIYKH VADFVEPESK SSSEPRSLTT LEIYAKAKEE QETTPVFSKK 360
361 TFKLEGSAIL VFVTKLSGKT KIHVATDFKE PVTLHWALSQ KGGEWLDPPS DILPPNSLPV 420
421 RGAVDTKLTI TSTDLPSPVQ TFELEIEGDS YKGMPFVLNA GERWIKNNDS DFYVDFAKEE 480
481 KHVQKDYGDG KGTAKHLLDK IADLESEAQK SFMHRFNIAA DLVDEAKSAG QLGFAGILVW 540
541 MRFMATRQLV WNKNYNVKPR EISKAQDRLT DLLQDVYASY PEYRELLRMI MSTVGRGGEG 600
601 DVGQRIRDEI LVIQRKNDCK GGIMEEWHQK LHNNTSPDDV VICQALMDYI KSDFDLSVYW 660
661 KTLNDNGITK ERLLSYDRAI HSEPNFRGEQ KDGLLRDLGH YMRTLKAVHS GADLESAIQN 720
721 CMGYQDDGEG FMVGVQINPV SGLPSGYPDL LRFVLEHVEE KNVEPLLEGL LEARQELRPL 780
781 LLKSHDRLKD LLFLDLALDS TVRTAIERGY EQLNDAGPEK IMYFISLVLE NLALSSDDNE 840
841 DLIYCLKGWQ FALDMCKSKK DHWALYAKSV LDRSRLALAS KAERYLEILQ PSAEYLGSCL 900
901 GVDQSAVSIF TEEIIRAGSA AALSSLVNRL DPVLRKTANL GSWQVISPVE VVGYVIVVDE 960
961 LLTVQNKTYD RPTIIVANRV RGEEEIPDGA VAVLTPDMPD VLSHVSVRAR NGKICFATCF1020
1021 DSGILSDLQG KDGKLLSLQP TSADVVYKEV NDSELSSPSS DNLEDAPPSI SLVKKQFAGR1080
1081 YAISSEEFTS DLVGAKSRNI GYLKGKVPSW VGIPTSVALP FGVFEKVISE KANQAVNDKL1140
1141 LVLKKTLDEG DQGALKEIRQ TLLGLVAPPE LVEELKSTMK SSDMPWPGDE GEQRWEQAWA1200
1201 AIKKVWASKW NERAYFSTRK VKLDHDYLCM AVLVQEVINA DYAFVIHTTN PSSGDSSEIY1260
1261 AEVVKGLGET LVGAYPGRSL SFICKKNNLD SPLVLGYPSK PIGLFIRRSI IFRSDSNGED1320
1321 LEGYAGAGLY DSVPMDEEDQ VVLDYTTDPL ITDLSFQKKV LSDIARAGDA IEKLYGTAQD1380
1381 IEGVIRDGKL YVVQTRPQV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
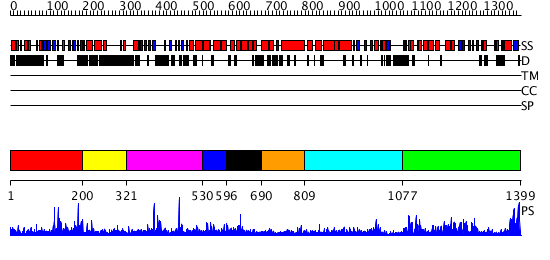
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [200..320] | 1.011996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [321..529] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [530..595] | 1.025995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [596..689] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [690..808] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [809..1076] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1077..1399] | 89.30103 | No description for 2olsA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)