













| General Information: |
|
| Name(s) found: |
gi|186893955
[NCBI NR]
gi|186696981 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Yersinia pseudotuberculosis PB1/+ |
| Length: | 819 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRVLKFGGTS VANAERFMRV ADIIESNARQ GQVATVLSAP AKITNHLVAM IDKMVAGQDI 60
61 SPNISDAERI FAELLRGLAD TQPGFDYDRL KALVGHEFAQ LKHLLHGISL LGQCPDSINA 120
121 SIICRGEKLS IAIMEALFQA KGYHVTVINP VEKLLAQGHY LESTVDITES TRRIGASGIP 180
181 SDHIILMAGF TAGNDKGELV VLGRNGSDYS AAVLAACLRA DCCEIWTDVD GVYTCDPRTV 240
241 PDARLLKSMS YQEAMELSYF GAKVLHPRTI APIARFQIPC LIKNTSNPQA PGTLIGGESI 300
301 DEDSPVKGIT NLNNMAMINV SGPGMKGMVG MAARVFAVMS RSGISVVLIT QSSSEYSISF 360
361 CVPQGELLRA RRALEDEFYL ELKDGVLDPL DVMEHLAIIS VVGDGMRTLR GISARFFSAL 420
421 ARANINIIAI AQGSSERSIS VVVNNDAVTT GVRVCHQMLF NTDQVIEVFV IGVGGVGGAL 480
481 IEQIYRQQPW LKQRHIDLRV CGIANSKAML TNVHGIALDN WRQELAEVQE PFNLSRLIRL 540
541 VKEYHLLNPV IVDCTSSQAV ADQYADFLTD GFHVVTPNKK ANTSSMNYYR QMRAAATKSC 600
601 RKFLYDTNVG AGLPVIENLQ NLLNAGDELM RFTGILSGSL SFIFGKLDEG MSLSEATRQA 660
661 KALGYTEPDP RDDLSGMDVA RKLLILAREA GYKLELADIE VESVLPASFD ASGDVDTFLA 720
721 RLPSLDAEFT RLVANAAEQG KVLRYVGVIE DGRCKVRMEA VDGNDPLYKV KNGENALAFY 780
781 TRYYQPIPLV LRGYGAGNDV TAAGVFADLL RTLSWKLGV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
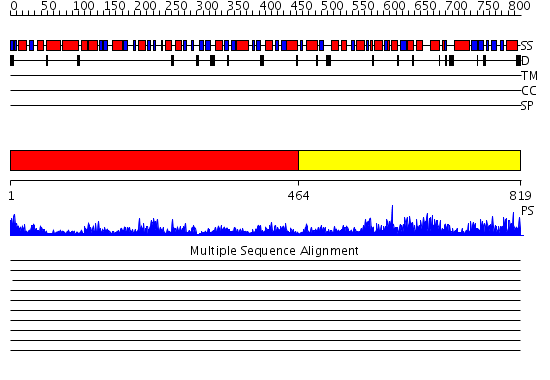
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..463] | 99.0 | No description for 2hmfA was found. | |
| 2 | View Details | [464..819] | 73.39794 | Homoserine Dehydrogenase in complex with suicide inhibitor complex NAD-5-hydroxy-4-Oxonorvaline |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)