













| General Information: |
|
| Name(s) found: |
ec-PA /
FBpp0100019
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1712 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
[NOT]protein deubiquitination
[IDA]
compound eye morphogenesis [IMP] retinal cell programmed cell death [TAS] compound eye retinal cell programmed cell death [TAS] ubiquitin-dependent protein catabolic process [IEA] |
| Molecular Function: |
[NOT]ubiquitin-specific protease activity
[IDA]
ubiquitin thiolesterase activity [IEA][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTAAIKTSL PHAPLAGQQQ PQQAQTTTAM AVAPTTITNA QTSAINAKNN IGAFFQTCKV 60
61 LWHLDAFRRS FRGLNQHVCG GQDCIFCALK ELFQQLQTSS EPALCPEPLR RALASGPLAG 120
121 RRFPLGCLGD AAECFELLLH RVHSHISPDD GDSCESSACI AHRRFAMRVI EQSVCKCGAN 180
181 SEQLPFTQMV HYVSASALTS QKSLALQSHQ QLSFGQLLRA AGNMGDIRDC PNTCGAKIGI 240
241 CRALLNRPEV VSIGIVWDSE RPAADQVHAV LKAVGTSLRL GDVFHQVSEP RWAQQTQHEL 300
301 VGIVSYYGKH YTTFFFHTKL KVWVYFDDAN VKEVGPSWEG VVDKCSRGRY QPLLLLYAVP 360
361 QQPTVGNGVG IGHVDVAMPA SAPSIAGSVV RRAVTPSPEK PSLGNTRRAI TPTPLRTPPN 420
421 DYQNLSVIQK NIFPSNLAMG TDETDAYISR KTVEHVLSAQ YQNLSVIQDK IGGGNGVGVG 480
481 QQSVDKDGGF LSRKPIEAVL SAQLARRQHL QLQRSHSAES SSGHASSNGS SPPSDGLTMP 540
541 EHLNQPRRRD SGNWSGDRNS ASSASSTTLD NPYLYMVAKR GVAAVPQSPT RHGLPYDPGY 600
601 DSFSLSSTDS YPPKHALNPQ LAKIPEAAMG VVPNGGAMPH GHGNGQLALS GDCEKLCHEA 660
661 DQLLEKSRLV EESHDLETAL VLCNAAAGRA RAAMDAPYSN PHTMTFARMK HNTCVMRARS 720
721 LHRRILVEKG AETEAMPELK HMREGSTSSV KHVRQNSKDR TLEKEQQQLQ FQQIQQIQQI 780
781 HQIQQQIPAS IEIYATLPKR KSPLKALASA SACDDNAIEY EQEKQLATSP GKPERESRSL 840
841 FGRKDKEKEK RSRSEDRNKL TREFSLTETL LVNAKDTLKK HKEDKDEKKE KDKSGKKQHK 900
901 IRRKLLMGGL IRRKNRSMPD LTEAVDDSTA NAGITHHPHY PAQGVTTAGT LLGTSVDDSA 960
961 VGLGKHGHGM SGYISEGHFD YSGSGSASNS GAGSNPNPNL ERSKLMRKSF HGSGRQLTMV1020
1021 PKVAPPPPMR TSSTLTPQQQ QQQQQQFHHE ANLSNLSAMS SNTSISEDSC QTIITTCAQV1080
1081 HSEQSPLKDL QMLVAQDELP LPPPMELPPY PSPPHSSCHS RQASEDFPPP PPPELDLEPL1140
1141 NQQLSQLQAL EAASKQQRQQ LESLEGTTSI LAQLQQRQHL LKLRKEQANH PGAATTDATW1200
1201 LKELQAKQAN DLRAMQRKLE ASSPSSVRDL THRFEQTSIR SYASQELLSQ PGQSQQLGQL1260
1261 RTQMNGLPNG NAKMDVDEVD AMPAVRNSLP AALSAHQLLP KPKYEMTQSQ IAEEIREVEL1320
1321 LNTMVQQTLN QNGAAALPKR VKKKSVSFCD QVILVATAGN EEDDDFIPNP ILERVLRTAQ1380
1381 HPNEKVTAQM IQQQQQNMMR LQTEAQPRQQ QVQQLQQPSP MLGRPAGATA DMQRYVQRMQ1440
1441 QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQHQELYRQ QQLHRTSMSG IASQQHLQQL1500
1501 DAQYTTMPRP LHHSQLGSYS VTLQKHQQLQ QQQQQQQQQQ APQQLSLGYF SNGSVATSEQ1560
1561 VPLPSPYQRV PLPHGYQPAA AGPYYPLPNA QQHLQQQQLM AKPVQKKVSF EPGTKGEADC1620
1621 LPPPPPPPQK NGLPETGSAG SGHPGAAPGA SGAGGTTSAG ITAIPTRVYN NAIVKASAKA1680
1681 VECNLCRKRH VIAPAVYCTN CEYYLQMLNQ RR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
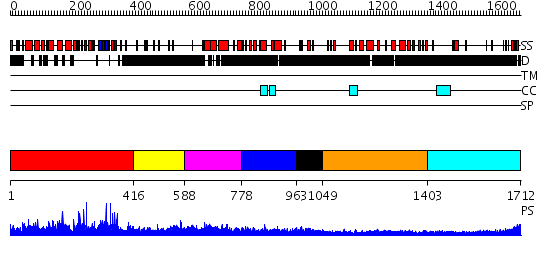
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..415] | 3.1 | Ubiquitin carboxyl-terminal hydrolase 7 (USP7, HAUSP) | |
| 2 | View Details | [416..587] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [588..777] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [778..962] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [963..1048] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1049..1402] | 17.221849 | Heavy meromyosin subfragment | |
| 7 | View Details | [1403..1712] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 |
|
||||||||||||
| 2 | No functions predicted. | ||||||||||||
| 3 | No functions predicted. | ||||||||||||
| 4 | No functions predicted. | ||||||||||||
| 5 | No functions predicted. | ||||||||||||
| 6 | No functions predicted. | ||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)