













| General Information: |
|
| Name(s) found: |
HEN1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 942 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] |
| Biological Process: |
virus induced gene silencing
[IMP]
specification of floral organ identity [IMP] regulation of flower development [IMP] production of miRNAs involved in gene silencing by miRNA [IMP][NAS] mRNA cleavage involved in gene silencing by miRNA [IMP] production of siRNA involved in RNA interference [IGI][IMP] |
| Molecular Function: |
RNA methyltransferase activity
[IDA][IMP]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGGGKHTPT PKAIIHQKFG AKASYTVEEV HDSSQSGCLG LAIPQKGPCL YRCHLQLPEF 60
61 SVVSNVFKKK KDSEQSAAEL ALDKLGIRPQ NDDLTVDEAR DEIVGRIKYI FSDEFLSAEH 120
121 PLGAHLRAAL RRDGERCGSV PVSVIATVDA KINSRCKIIN PSVESDPFLA ISYVMKAAAK 180
181 LADYIVASPH GLRRKNAYPS EIVEALATHV SDSLHSREVA AVYIPCIDEE VVELDTLYIS 240
241 SNRHYLDSIA ERLGLKDGNQ VMISRMFGKA SCGSECRLYS EIPKKYLDNS SDASGTSNED 300
301 SSHIVKSRNA RASYICGQDI HGDAILASVG YRWKSDDLDY DDVTVNSFYR ICCGMSPNGI 360
361 YKISRQAVIA AQLPFAFTTK SNWRGPLPRE ILGLFCHQHR LAEPILSSST APVKSLSDIF 420
421 RSHKKLKVSG VDDANENLSR QKEDTPGLGH GFRCEVKIFT KSQDLVLECS PRKFYEKEND 480
481 AIQNASLKAL LWFSKFFADL DVDGEQSCDT DDDQDTKSSS PNVFAAPPIL QKEHSSESKN 540
541 TNVLSAEKRV QSITNGSVVS ICYSLSLAVD PEYSSDGESP REDNESNEEM ESEYSANCES 600
601 SVELIESNEE IEFEVGTGSM NPHIESEVTQ MTVGEYASFR MTPPDAAEAL ILAVGSDTVR 660
661 IRSLLSERPC LNYNILLLGV KGPSEERMEA AFFKPPLSKQ RVEYALKHIR ESSASTLVDF 720
721 GCGSGSLLDS LLDYPTSLQT IIGVDISPKG LARAAKMLHV KLNKEACNVK SATLYDGSIL 780
781 EFDSRLHDVD IGTCLEVIEH MEEDQACEFG EKVLSLFHPK LLIVSTPNYE FNTILQRSTP 840
841 ETQEENNSEP QLPKFRNHDH KFEWTREQFN QWASKLGKRH NYSVEFSGVG GSGEVEPGFA 900
901 SQIAIFRREA SSVENVAESS MQPYKVIWEW KKEDVEKKKT DL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
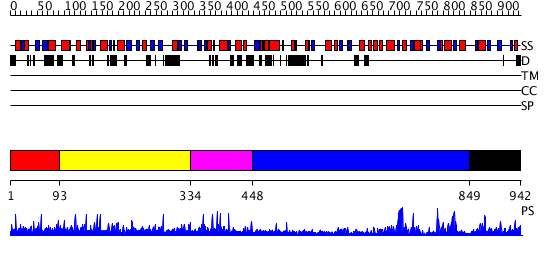
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [93..333] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [334..447] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [448..848] | 14.69897 | Crystal Structure of RumA, the iron-sulfur cluster containing E. coli 23S Ribosomal RNA 5-Methyluridine Methyltransferase | |
| 5 | View Details | [849..942] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)