













| General Information: |
|
| Name(s) found: |
gi|79536755
[NCBI NR]
gi|56381941 [NCBI NR] gi|44917469 [NCBI NR] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 726 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
protein folding
[IEA][ISS]
|
| Molecular Function: |
heat shock protein binding
[IEA]
unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MECNKDEAKR AMDIAERKMT EKDYTGAKKF ANKAQNLFPE LDGLKQLFVA INVYISGEKT 60
61 FAGEADWYGV LGVDPFASDE ALKKQYRKLV LMLHPDKNKC KGAEGAFNLV AEAWALLSDK 120
121 DKRILYNVKR GKDVKAAQQR FPTTQREIPS HQPTSNGIPN VREHVVSSAR ARYKPATRKP 180
181 AARMDRSRTG SPAFVYPTQE SSTFWTMCNK CDTQYEYQRV YLNQTLLCPH CHHGFVAEEK 240
241 TPPTNIPKPP VNISSNQHHR SSKNQASNKN SNGSSYRREP ATSVNHNFQW DSSRMGGSYS 300
301 RNATNETANV VQQGQDKLKR VFWETQEREA ARGFTNSDLG NFKRQKTDDS HMRGPSAGSR 360
361 HPYVQALLRS DIKKALMDRG QSEIFKRLPM MIAKMEGKVN PTEGEKNSTK AMSSKASEVE 420
421 RSKMSSTANE VERSVEVIPH ESDEVKEIVV PDSDFHNFDL DRSESAFKDD QIWAAYDDAD 480
481 GMPRFYARIQ KVISVNPFKL KISWLNSKTT SEFGPIDWMG AGFAKSCGDF RCGRYESTDT 540
541 LNAFSHSVDF TKGARGLLHI LPKKGQVWAL YRNWSPEWDK NTPDEVKHKY EMVEVLDDYT 600
601 EDDQSLTVAL LLKAEGFRVV FRRCTEKLGV RKIAKEEMLR FSHQVPHYIL TGKEADNAPE 660
661 GFLELDPAAT PCAFSSENAE ADEKSEAVKE NEQGEAVKEN EESEALKENE ESEAVKENEE 720
721 VGMDLD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
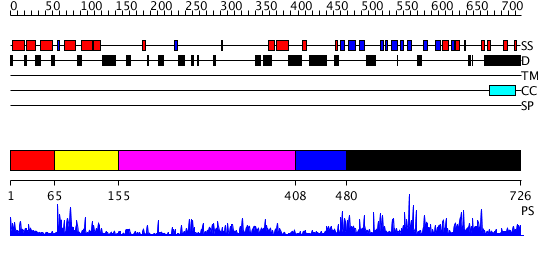
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..64] | 4.09691 | Designed protein cTPR2 | |
| 2 | View Details | [65..154] | 20.0 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 3 | View Details | [155..407] | 29.09691 | The crystal structure of Hsp40 Ydj1 | |
| 4 | View Details | [408..479] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [480..726] | 2.039952 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)