













| General Information: |
|
| Name(s) found: |
XRN3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1020 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
miRNA catabolic process
[IMP]
|
| Molecular Function: |
5'-3' exoribonuclease activity
[IGI][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVPSFYRWL AEKYPLLVAD VIEEEPVEIE GIKIPVDTSK PNPNNLEYDN LYLDMNGIIH 60
61 PCFHPEDRPS PTTFEEVFQC MFDYIDRLFV MVRPRKLLYM AIDGVAPRAK MNQQRSRRFR 120
121 SAKDASDAAA EEERLREEFE REGRRLPPKV DSQVFDSNVI TPGTEFMGVL SIALQYYVHL 180
181 RLNHDVGWKN IKVILSDANV PGEGEHKIMS YIRLQRNLPG FDPNTRHCLY GLDADLIMLG 240
241 LATHEVHFSI LREVVYTPGQ QERCFLCGQM GHFASNCEGK PKKRAGESDE KGDGNDFVKK 300
301 PYQFLHIWVL REYLELEMRI PNPPFEIDLE RIVDDFIFIC FFVGNDFLPH MPTLEIREGA 360
361 INLLMAVYKK EFRSFDGYLT DGCKPNLKRV EQFIQAVGSF EDKIFQKRAM QHQRQAERVK 420
421 RDKAGKATKR MDDEAPTVQP DLVPVARFSG SRLASAPTPS PFQSNDGRSA PHQKVRRLSP 480
481 GSSVGAAIVD VENSLESDER ENKEELKTKL KELIREKSDA FNSDTTEEDK VKLGQPGWRE 540
541 RYYEEKFSVV TPEEMERVRK DVVLKYTEGL CWVMHYYMEG VCSWQWFYPY HYAPFASDLK 600
601 DLGEMDIKFE LGTPFKPFNQ LLGVFPAASS HALPERYRTL MTDPNSPIID FYPTDFEVDM 660
661 NGKRFSWQGI AKLPFIDERR LLEAVSEVEF TLTDEEKRRN SRMCDMLFIA TSHRLAELVF 720
721 SLDNHCRQLS ARERVDFKVK IKPKLSDGMN GYLTPCSGET HPPVFRSPME GMEDILTNQV 780
781 ICCIYRLPDA HEHITRPPPG VIFPKKTVDI GDLKPPPALW HEDNGRRPMH NNHGMHNNHG 840
841 MHNNQGRQNP PGSVSGRHLG NAAHRLVSNS LQMGTDRYQT PTDVPAPGYG YNPPQYVPPI 900
901 PYQHGGYMAP PGAQGYAQPA PYQNRGGYQP RGPSGRFPSE PYQSQSREGQ HASRGGGYSG 960
961 NHQNQHQQQQ WHGQGGSEQN NPRGYNGQHH HQQGGDHDRR GRGRGSHHHH DQGGNPRHRY1020
1021 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
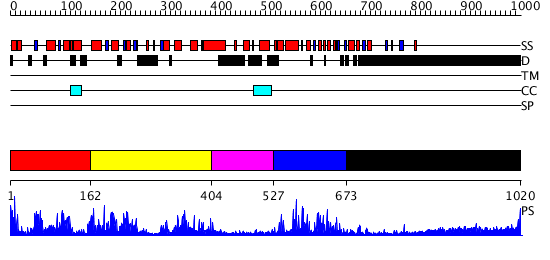
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 3.108967 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [162..403] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [404..526] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [527..672] | 1.093948 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [673..1020] | 25.221849 | No description for 1y0fB was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)