













| General Information: |
|
| Name(s) found: |
TPS8_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 856 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
metabolic process
[IEA]
trehalose biosynthetic process [IEA] |
| Molecular Function: |
transferase activity, transferring glycosyl groups
[ISS]
[NOT]alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity [IGI][IMP] [NOT]trehalose-phosphatase activity [IGI][IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSRSCANFL DLSSWDLLDF PQTPRTLPRV MTVPGIITDV DGDTTSEVTS TSGGSRERKI 60
61 IVANMLPLQS KRDAETGKWC FNWDEDSLQL QLRDGFSSET EFLYVGSLNV DIETNEQEEV 120
121 SQKLLEEFNC VATFLSQELQ EMFYLGFCKH QLWPLFHYML PMFPDHGDRF DRRLWQAYVS 180
181 ANKIFSDRVM EVINPEDDYV WIQDYHLMVL PTFLRKRFNR IKLGFFLHSP FPSSEIYRTL 240
241 PVRDEILRGL LNCDLIGFHT FDYARHFLSC CSRMLGLDYE SKRGHIGLDY FGRTVYIKIL 300
301 PVGVHMGRLE SVLSLDSTAA KTKEIQEQFK GKKLVLGIDD MDIFKGISLK LIAMEHLFET 360
361 YWHLKGKVVL VQIVNPARSS GKDVEEAKRE TYETARRINE RYGTSDYKPI VLIDRLVPRS 420
421 EKTAYYAAAD CCLVNAVRDG MNLVPYKYIV CRQGTRSNKA VVDSSPRTST LVVSEFIGCS 480
481 PSLSGAIRVN PWDVDAVAEA VNSALKMSET EKQLRHEKHY HYISTHDVGY WAKSFMQDLE 540
541 RACRDHYSKR CWGIGFGLGF RVLSLSPSFR KLSVEHIVPV YRKTQRRAIF LDYDGTLVPE 600
601 SSIVQDPSNE VVSVLKALCE DPNNTVFIVS GRGRESLSNW LSPCENLGIA AEHGYFIRWK 660
661 SKDEWETCYS PTDTEWRSMV EPVMRSYMEA TDGTSIEFKE SALVWHHQDA DPDFGSCQAK 720
721 EMLDHLESVL ANEPVVVKRG QHIVEVKPQG VSKGLAAEKV IREMVERGEP PEMVMCIGDD 780
781 RSDEDMFESI LSTVTNPELL VQPEVFACTV GRKPSKAKYF LDDEADVLKL LRGLGDSSSS 840
841 LKPSSSHTQV AFESIV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
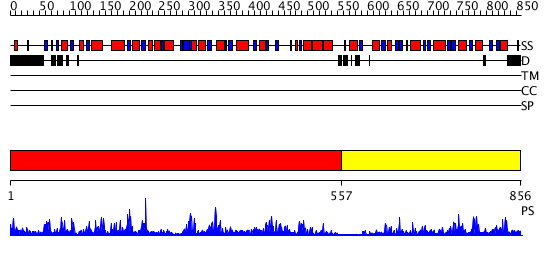
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..556] | 81.69897 | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | |
| 2 | View Details | [557..856] | 26.0 | Crystal structure of NYSGRC target T1436: A Hypothetical protein yidA. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)