













| General Information: |
|
| Name(s) found: |
TPS6_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 860 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
|
| Biological Process: |
trehalose biosynthetic process
[IGI][ISS]
|
| Molecular Function: |
transferase activity, transferring glycosyl groups
[ISS]
trehalose-phosphatase activity [IGI][IMP][ISS] alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity [IGI][IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSRSYSNLL ELASGDSPTF GRMNRQIPRI MAVAGIMSNI DNDSKDTDLS PKDRIIIVAN 60
61 ELPIRAQRRV DGNGSGSSSS STCCSKGWNF SWDENSLLLQ LKDGLGDEAI EVIYVGCLKE 120
121 EIPLNEQEEV YQILLESFKC VPTFLPLDLY TRYYHGFCKQ QLWPLFHYML PLSPDLGGRF 180
181 DRTLWQAYVS VNKIFADRIM EVINPEDDFV WIHDYHLMVL PTFLRKRFNR VKLGFFLHSP 240
241 FPSSEIYKTL PIREELLRAL LNSDLIGFHT FDYARHFLSC CSRMLGLTYE SKRGYIGLEY 300
301 YGRTVSIKIL PVGIHMGQLQ SVLSLPETER KVGELIERYG RKGRTMLLGV DDMDIFKGIT 360
361 LKLLAMEQLL MQHPEWQGKV VLVQIANPAR GKGKDVKEMQ AETYSTVKRI NETFGRPGYD 420
421 PIVLIDAPLK FYERVAYYVV AECCLVTAVR DGMNLIPYEY IVSRQGNEKL DKILKLEANN 480
481 RNKKSMLVVS EFIGCSPSLS GAIRVNPWNV DAVADAMDSA LEVAEPEKQL RHEKHYKYVS 540
541 THDVGYWARS FLQDLERSCG EHGRRRCWGI GFGLSFRVVA LDQSFRKLSM EHIVSAYKRT 600
601 KTRAILLDYD DTLMPQGSID KRPSSKSIDI LNTLCRDKGN LVFIVSAKSR ETLSDWFSPC 660
661 EKLGIAAEHG YFLRLRKAVE WENCVAAVDC SWKQIAEPVM ELYTETTDGS TIEDKETALV 720
721 WSYEDADPDF GSCQAKELLD HLESVLANEP VTVKRGQNYV EVKPQGVSKG LIARRMLSMM 780
781 QERGTLPEFV LCIGDDRSDE DMFEVICSST EGPSIAPRAE IFACTVGQKP SKAKYYLDDT 840
841 TEIVRLMHGL ASVTDQITPV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
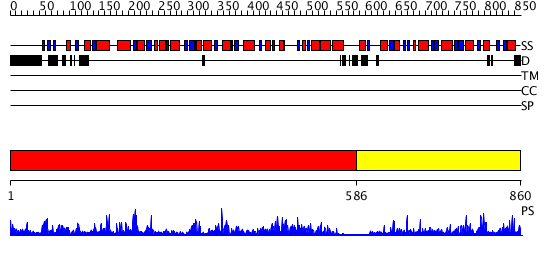
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..585] | 78.30103 | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | |
| 2 | View Details | [586..860] | 23.39794 | Crystal structure of NYSGRC target T1436: A Hypothetical protein yidA. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)