













| General Information: |
|
| Name(s) found: |
NU98B_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 997 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFGSSNNNPF GQSSISSPFG TQTHSLFGQT NNNASNNPFA TKPFGTSTPF GAQTGSSMFG 60
61 GTSTGVFGAP QTSSPFGASP QAFGSSTQAF GASSTPSFGS SNSPFGGTST FGQKSFGLST 120
121 PQSSPFGSTT QQSQPAFGNS TFGSSTPFGA STTPAFGASS TPAFGVSNTS GFGATNTPGF 180
181 GATNTTGFGG SSTPGFGASS TPAFGSTNTP AFGASSTPLF GSSSSPAFGA SPAPAFGSSG 240
241 NAFGNNTFSS GGAFGSSSTP TFGASNTSAF GASSSPSFNF GSSPAFGQST SAFGSSSFGS 300
301 TQSSLGSTPS PFGAQGAQAS TSTFGGQSTI GGQQGGSRVI PYAPTTDTAS GTESKSERLQ 360
361 SISAMPAHKG KNMEELRWED YQRGDKGGQR STGQSPEGAG FGVTNSQPSI FSTSPAFSQT 420
421 PVNPTNPFSQ TTPTSNTNFS PSFSQPTTPS FGQPTTPSFR STVSNTTSVF GSSSSLTTNT 480
481 SQPLGSSIFG STPAHGSTPG FSIGGFNNSQ SSPLFGSNPS FAQNTTPAFS QTSPLFGQNT 540
541 TPALGQSSSV FGQNTNPALV QSNTFSTPST GFGNTFSSSS SLTTSISPFG QITPAVTPFQ 600
601 SAQPTQPLGA FGFNNFGQTQ IANTTDIAGA MGTFSQGNFK QQPALGNSAV MQPTPVTNPF 660
661 GTLPALPQIS IAQGGNSPSI QYGISSMPVV DKPAPVRVSP LLTSRHLLQR RVRLPTRKYR 720
721 PSDDGPKVPF FSDEEENSST PKADAFFIPR ENPRALFIRP VERVKSEHPK DSPTPLQENG 780
781 KRSNGVTNGA NHETKDNGAI REAPPVKVNQ KQNGTHENHG GDKNGSHSSP SGADIESLMP 840
841 KLHHSEYFTE PRIQELAAKE RVEQGYCKRV KDFVVGRHGY GSIKFLGETD VCRLDLEMVV 900
901 QFKNREVNVY MDESKKPPVG QGLNKPAVVT LLNIKCMDKK TGTQVMEGER LDKYKEMLKR 960
961 KAGEQGAQFV SYDPVNGEWT FKVEHFSSYK LGDEYDV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
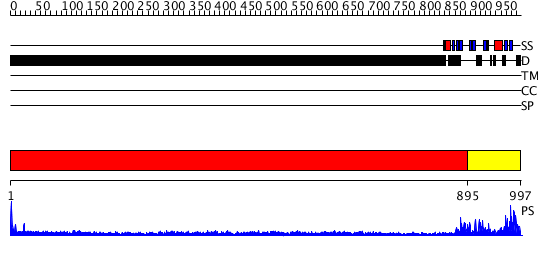
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..894] | 34.30103 | No description for 1y0fB was found. | |
| 2 | View Details | [895..997] | 52.30103 | C-terminal autoproteolytic domain of nucleoporin nup98 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)