













| General Information: |
|
| Name(s) found: |
Y1044_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 665 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
response to light stimulus
[IEA][ISS]
|
| Molecular Function: |
protein binding
[IEA]
signal transducer activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MACMKLGSKS DAFQRQGQAW FCTTGLPSDI VVEVGEMSFH LHKFPLLSRS GVMERRIAEA 60
61 SKEGDDKCLI EISDLPGGDK TFELVAKFCY GVKLELTASN VVYLRCAAEH LEMTEEHGEG 120
121 NLISQTETFF NQVVLKSWKD SIKALHSCDE VLEYADELNI TKKCIESLAM RASTDPNLFG 180
181 WPVVEHGGPM QSPGGSVLWN GISTGARPKH TSSDWWYEDA SMLSFPLFKR LITVMESRGI 240
241 REDIIAGSLT YYTRKHLPGL KRRRGGPESS GRFSTPLGSG NVLSEEEQKN LLEEIQELLR 300
301 MQKGLVPTKF FVDMLRIAKI LKASPDCIAN LEKRIGMQLD QAALEDLVMP SFSHTMETLY 360
361 DVDSVQRILD HFLGTDQIMP GGVGSPCSSV DDGNLIGSPQ SITPMTAVAK LIDGYLAEVA 420
421 PDVNLKLPKF QALAASIPEY ARLLDDGLYR AIDIYLKHHP WLAETERENL CRLLDCQKLS 480
481 LEACTHAAQN ERLPLRIIVQ VLFFEQLQLR TSVAGCFLVS DNLDGGSRQL RSGGYVGGPN 540
541 EGGGGGGGWA TAVRENQVLK VGMDSMRMRV CELEKECSNM RQEIEKLGKT TKGGGSASNG 600
601 VGSKTWENVS KKLGFGFKLK SHQMCSAQEG SVSKSNNENV KIEKLKDVKE RRGKHKKASS 660
661 ISSER |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
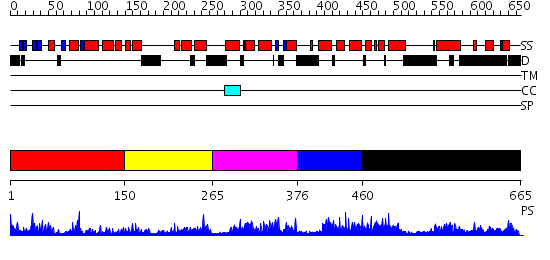
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..149] | 19.39794 | No description for 2nn2A was found. | |
| 2 | View Details | [150..264] | 1.645994 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [265..375] | 1.098984 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [376..459] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [460..665] | 3.077978 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)