













| General Information: |
|
| Name(s) found: |
DNAJ1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 408 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[ISS]
|
| Biological Process: |
protein folding
[IEA][ISS]
|
| Molecular Function: |
nucleic acid binding
[IEA]
heat shock protein binding [IEA] zinc ion binding [IEA] unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRRFNWVLRH VQARRTFDSA IGLRQGSQKP LFERYIHATG INNSSARNYY DVLGVSPKAT 60
61 REEIKKSFHE LAKKFHPDTN RNNPSAKRKF QEIREAYETL GNSERREEYD KLQYRNSDYV 120
121 NNDGGDSERF RRAYQSNFSD TFHKIFSEIF ENNQIKPDIR VELSLSLSEA AEGCTKRLSF 180
181 DAYVFCDSCD GLGHPSDAAM SICPTCRGVG RVTIPPFTAS CQTCKGTGHI IKEYCMSCRG 240
241 SGIVEGTKTA ELVIPGGVES EATITIVGAG NVSSRTSQPG NLYIKLKVAN DSTFTRDGSD 300
301 IYVDANISFT QAILGGKVVV PTLSGKIQLD IPKGTQPDQL LVLRGKGLPK QGFFVDHGDQ 360
361 YVRFRVNFPT EVNERQRAIL EEFAKEEINN ELSDSAEGSW LYQKLSTG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
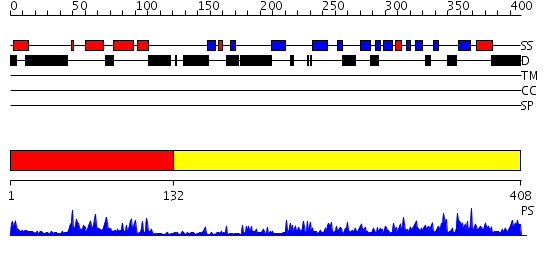
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | 26.154902 | J-DOMAIN (RESIDUES 1-77) OF THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 1-104) OF THE MOLECULAR CHAPERONE DNAJ, NMR, 20 STRUCTURES | |
| 2 | View Details | [132..408] | 48.0 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)