













| General Information: |
|
| Name(s) found: |
F26_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 744 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA]
plasma membrane [IDA] |
| Biological Process: |
fructose metabolic process
[IMP]
|
| Molecular Function: |
fructose-2,6-bisphosphate 2-phosphatase activity
[IMP]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSGASKNTE EDDDGSNGGG GQLYVSLKME NSKVEGELTP HVYGSLPLIG SWDPSKALPM 60
61 QRESALMSEL SFVVPPDHET LDFKFLLKPK NRNTPCIVEE GENRLLTGGS LQGDARLALF 120
121 RLEGDVIVEF RVFINADRVS PIDLATSWRA YRENLQPSTV RGIPDVSINP DPKSAECPLE 180
181 SLELDLAHYE VPAPAPSANS YLVYAADNAE NPRSLSASGS FRNDSTPKAA QRNSEDSGVT 240
241 VDGSPSAKEM TIVVPDSSNI YSAFGEAESK SVETLSPFQQ KDGQKGLFVD RGVGSPRLVK 300
301 SLSASSFLID TKQIKNSMPA AAGAVAAAAV ADQMLGPKED RHLAIVLVGL PARGKTFTAA 360
361 KLTRYLRWLG HDTKHFNVGK YRRLKHGVNM SADFFRADNP EGVEARTEVA ALAMEDMIAW 420
421 MQEGGQVGIF DATNSTRVRR NMLMKMAEGK CKIIFLETLC NDERIIERNI RLKIQQSPDY 480
481 SEEMDFEAGV RDFRDRLANY EKVYEPVEEG SYIKMIDMVS GNGGQIQVNN ISGYLPGRIV 540
541 FFLVNTHLTP RPILLTRHGE SMDNVRGRIG GDSVISDSGK LYAKKLASFV EKRLKSEKAA 600
601 SIWTSTLQRT NLTASSIVGF PKVQWRALDE INAGVCDGMT YEEVKKNMPE EYESRKKDKL 660
661 RYRYPRGESY LDVIQRLEPV IIELERQRAP VVVISHQAVL RALYAYFADR PLKEIPQIEM 720
721 PLHTIIEIQM GVSGVQEKRY KLMD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
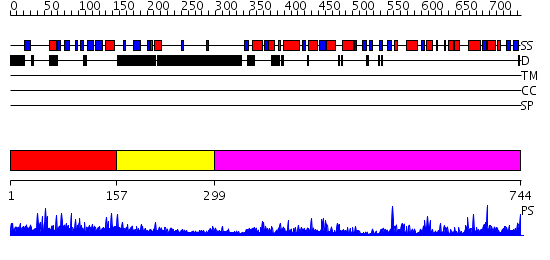
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..156] | 4.73 | beta-amylase | |
| 2 | View Details | [157..298] | 4.39794 | Crystal Structure of a GTP-Regulated ATP Sulfurylase Heterodimer from Pseudomonas syringae | |
| 3 | View Details | [299..744] | 126.0 | Crystal structure of the human inducible form 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)