













| General Information: |
|
| Name(s) found: |
gi|42562111
[NCBI NR]
gi|209529769 [NCBI NR] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 641 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
signal transduction
[IEA]
regulation of transcription, DNA-dependent [IEA] |
| Molecular Function: |
transcription factor activity
[IEA]
protein binding [IEA] signal transducer activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGDCAIDTE KYSLLEDFNV DVEVENKAFE TFSLCFWVYL LDSTTYPSAI IRQVHSDMSV 60
61 SAPFLVLDEN KKMMLLPLTL LHREAPDPVN TSSWTEVPNV STTAKFPLQK WVHVGCEVSR 120
121 NYMRLYICGE LVGEQVLTSL MTNGTNSDCA RKISLFSVGG DGYSVQGFIH SAEVLPSNLS 180
181 ASYHYTKDPP LWLSVDKPST SGIELDEDGV WSVVSGTFCS LDVVLTNAIG QPVHKDVKVV 240
241 ASLLYADSGT HVEKRSDFEA FLLVSYEGIE LSAEDKPCNL LNGCASFKFK LSQLSSKSDK 300
301 RLFCIKFEIP EVKANYPFLE TVTNQIRCIS RNRDSVSSMK RIRLGEEKVS ESKIVNGNGT 360
361 SMEWRPQNHE EDNSSTDSEN TEMRDSTAFR RYSIPDWIIF KYCLGNLTER ALLLKEITNN 420
421 SSDEEVSEFA DQVSLYSGCS HHGYQIKMAR KLIAEGTNAW NLISRNYRHV HWDNVVIEIE 480
481 EHFMRIAKCS SRSLTHQDFD LLRRICGCYE YITQENFETM WCWLFPVASA VSRGLINGMW 540
541 RSASPKWIEG FVTKEEAERS LQNQVPGTFI LRFPTSRSWP HPDAGSVVVT YVGHDLVIHH 600
601 RLLTINHICD SSERYTDAKQ LQDMLLAEPE LSRLGRIIRG I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
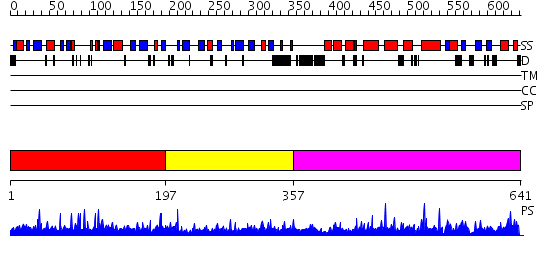
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | 1.15 | No description for 2uurA was found. | |
| 2 | View Details | [197..356] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [357..641] | 98.69897 | STAT3b |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)