













| General Information: |
|
| Name(s) found: |
HAC12_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1706 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
|
| Biological Process: |
flower development
[IGI]
regulation of transcription, DNA-dependent [ISS] protein amino acid acetylation [IMP] |
| Molecular Function: |
histone acetyltransferase activity
[IDA][ISS]
transcription cofactor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNVQAHMSGQ RSGQVPNQGT VPQNNGNSQM QNLVGSNGAA TAVTGAGAAT GSGTGVRPSR 60
61 NIVGAMDHDI MKLRQYMQTL VFNMLQQRQP SPADAASKAK YMDVARRLEE GLFKMAVTKE 120
121 DYMNRSTLES RITSLIKGRQ INNYNQRHAN SSSVGTMIPT PGLSQTAGNP NLMVTSSVDA 180
181 TIVGNTNITS TALNTGNPLI AGGMHGGNMS NGYQHSSRNF SLGSGGSMTS MGAQRSTAQM 240
241 IPTPGFVNSV TNNNSGGFSA EPTIVPQSQQ QQQRQHTGGQ NSHMLSNHMA AGVRPDMQSK 300
301 PSGAANSSVN GDVGANEKIV DSGSSYTNAS KKLQQGNFSL LSFCPDDLIS GQHIESTFHI 360
361 SGEGYSTTNP DPFDGAITSA GTGTKAHNIN TASFQPVSRV NSSLSHQQQF QQPPNRFQQQ 420
421 PNQIQQQQQQ FLNQRKLKQQ TPQQHRLISN DGLGKTQVDS DMVTKVKCEP GMENKSQAPQ 480
481 SQASERFQLS QLQNQYQNSG EDCQADAQLL PVESQSDICT SLPQNSQQIQ QMMHPQNIGS 540
541 DSSNSFSNLA VGVKSESSPQ GQWPSKSQEN TLMSNAISSG KHIQEDFRQR ITGMDEAQPN 600
601 NLTEGSVIGQ NHTSTISESH NLQNSIGTTC RYGNVSHDPK FKNQQRWLLF LRHARSCKPP 660
661 GGRCQDQNCV TVQKLWSHMD NCADPQCLYP RCRHTKALIG HYKNCKDPRC PVCVPVKTYQ 720
721 QQANVRALAR LKNESSAVGS VNRSVVSNDS LSANAGAVSG TPRCADTLDN LQPSLKRLKV 780
781 EQSFQPVVPK TESCKSSIVS TTEADLSQDA ERKDHRPLKS ETMEVKVEIP DNSVQAGFGI 840
841 KETKSEPFEN VPKPKPVSEP GKHGLSGDSP KQENIKMKKE PGWPKKEPGC PKKEELVESP 900
901 ELTSKSRKPK IKGVSLTELF TPEQVREHIR GLRQWVGQSK AKAEKNQAME NSMSENSCQL 960
961 CAVEKLTFEP PPIYCTPCGA RIKRNAMYYT VGGGETRHYF CIPCYNESRG DTILAEGTSM1020
1021 PKAKLEKKKN DEEIEESWVQ CDKCQAWQHQ ICALFNGRRN DGGQAEYTCP YCYVIDVEQN1080
1081 ERKPLLQSAV LGAKDLPRTI LSDHIEQRLF KRLKQERTER ARVQGTSYDE IPTVESLVVR1140
1141 VVSSVDKKLE VKSRFLEIFR EDNFPTEFPY KSKVVLLFQK IEGVEVCLFG MYVQEFGSEC1200
1201 SNPNQRRVYL SYLDSVKYFR PDIKSANGEA LRTFVYHEIL IGYLEYCKLR GFTSCYIWAC1260
1261 PPLKGEDYIL YCHPEIQKTP KSDKLREWYL AMLRKAAKEG IVAETTNLYD HFFLQTGECR1320
1321 AKVTAARLPY FDGDYWPGAA EDIISQMSQE DDGRKGNKKG ILKKPITKRA LKASGQSDFS1380
1381 GNASKDLLLM HKLGETIHPM KEDFIMVHLQ HSCTHCCTLM VTGNRWVCSQ CKDFQLCDGC1440
1441 YEAEQKREDR ERHPVNQKDK HNIFPVEIAD IPTDTKDRDE ILESEFFDTR QAFLSLCQGN1500
1501 HYQYDTLRRA KHSSMMVLYH LHNPTAPAFV TTCNVCHLDI ESGLGWRCEV CPDYDVCNAC1560
1561 YKKEGCINHP HKLTTHPSLA DQNAQNKEAR QLRVLQLRKM LDLLVHASQC RSPVCLYPNC1620
1621 RKVKGLFRHG LRCKVRASGG CVLCKKMWYL LQLHARACKE SECDVPRCGD LKEHLRRLQQ1680
1681 QSDSRRRAAV MEMMRQRAAE VAGTSG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
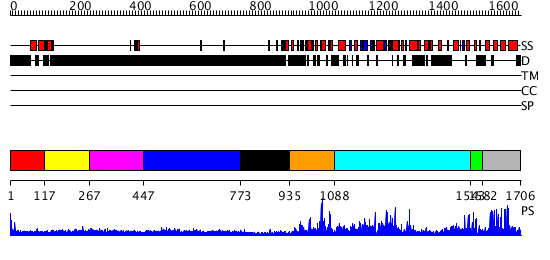
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [117..266] | 7.0 | Sec24 | |
| 3 | View Details | [267..446] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [447..772] | 1.223995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [773..934] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [935..1087] | 31.69897 | No description for 2i13A was found. | |
| 7 | View Details | [1088..1542] | 136.0 | No description for 3biyA was found. | |
| 8 | View Details | [1543..1581] | 17.0 | ZZ Domain of CBP- a Novel Fold for a Protein Interaction Module | |
| 9 | View Details | [1582..1706] | 27.522879 | CREB-binding transcriptional adaptor protein CBP (p300) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)