













| General Information: |
|
| Name(s) found: |
STR2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 342 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
cytosol [IEP] |
| Biological Process: |
sulfate transport
[IEA]
aging [ISS] |
| Molecular Function: |
thiosulfate sulfurtransferase activity
[ISS]
3-mercaptopyruvate sulfurtransferase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRAFSSQLR SAYPASKSTH FGRVMASSGS EAKANYAPIS TNEPVVSVDW LHSNLGDADI 60
61 KVLDASWYMA HEQRNPIQEY QVAHIPGALF FDLNGIADRK TNLRHMLPSE EAFAAGCSAL 120
121 GIENNDGVVV YDGMGLFSAA RVWWMFRVFG HDKVWVLDGG LPKWRASGYD VESSVSNDAI 180
181 LKASAATEAI EKIYQGQTIS PITFQTKFRP HLVLALDQVK ENIEDKTYQH IDARSKARFD 240
241 GIAPEPWKGL PSGHIPGSKC VPFPLMFDSS QTLLPAEELK KQFEQEDISL DSPIAASCGT 300
301 GVTACILALG LYRLGKTNVA IYDGSWTEWA TAPNLPIVGS SS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
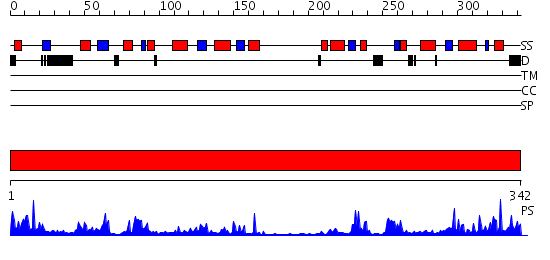
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..342] | 64.522879 | The "Rhodanese" fold and catalytic mechanism of 3-mercaptopyruvate sulfotransferases: Crystal structure of SseA from Escherichia coli |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.48 |
Source: Reynolds et al. (2008)