













| General Information: |
|
| Name(s) found: |
P4KB2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1116 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
pollen tube growth
[IGI]
root hair cell tip growth [IGI] phosphoinositide biosynthetic process [TAS] |
| Molecular Function: |
inositol or phosphatidylinositol kinase activity
[IEA]
binding [IEA] phosphotransferase activity, alcohol group as acceptor [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQMAQFLSLV RGDSIESPRE ITSPSNLISE SGSNGWLIRF FDSSFFCEWI AVSYLYKHQH 60
61 SGVRDYLCNR MYTLPLSGIE SYLFQICYLM VHKPSPSLDK FVIDICAKSL KIALKVHWFL 120
121 LAELEDSDDN EGISRIQEKC QIAATLVGEW SPLMRPHNEP STPGSKVLNK FLSSKQKLFS 180
181 LTLSPPTQKS LLFSPTSGSN LQDDGSQLSA DDNKIFKRLI PSPKVRDALL FRKSADKEDE 240
241 ECEKDGFFKR LLRDSRGEDD EQRSNSEGFF KRLLKDNKSE EEEISNNSEG FFKRLRSSKG 300
301 DEEELTSSSD GFFKRLLRDN KGDEEELGAN SEGFFKKLLR DSKNEDEEPN ANTEGFFKKL 360
361 FHESKNEDDK VSNAVDDEEK DGFLKKLFKE KFDEKRNGNE RNETDETVYT DETSGEDNGR 420
421 EGFFKKLFKE KFEDKPNIGK ADDGNESEDD ESSEFSLFRR LFRRHPEDVK TTLPSENCSN 480
481 GGFVESSPGT ENFFRKLFRD RDRSVEDSEL FGSKKYKEKC PGSPKPQNNT PSKKPPLPNN 540
541 TAAQFRKGSY HESLEFVHAL CETSYDLVDI FPIEDRKTAL RESIAEINSH LAQAETTGGI 600
601 CFPMGRGVYR VVNIPEDEYV LLNSREKVPY MICVEVLKAE TPCGAKTTST SLKLSKGGIP 660
661 LANGDAFLHK PPPWAYPLST AQEVYRNSAD RMSLSTVEAI DQAMTHKSEV KLVNACLSVE 720
721 THSNSNTKSV SSGVTGVLRT GLESDLEWVR LVLTADPGLR MESITDPKTP RRKEHRRVSS 780
781 IVAYEEVRAA AAKGEAPPGL PLKGAGQDSS DAQPMANGGM LKAGDALSGE FWEGKRLRIR 840
841 KDSIYGNLPG WDLRSIIVKS GDDCRQEHLA VQLISHFFDI FQEAGLPLWL RPYEVLVTSS 900
901 YTALIETIPD TASIHSIKSR YPNITSLRDF FDAKFKENSP SFKLAQRNFV ESMAGYSLVC 960
961 YLLQIKDRHN GNLLMDEEGH IIHIDFGFML SNSPGGVNFE SAPFKLTREL LEVMDSDAEG1020
1021 LPSEFFDYFK VLCIQGFLTC RKHAERIILL VEMLQDSGFP CFKGGPRTIQ NLRKRFHLSL1080
1081 TEEQCVSLVL SLISSSLDAW RTRQYDYYQR VLNGIR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
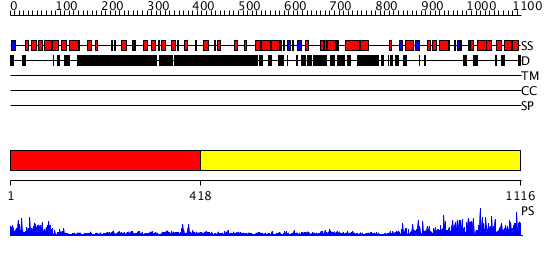
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..417] | 10.045757 | Heavy meromyosin subfragment | |
| 2 | View Details | [418..1116] | 78.0 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)