













| General Information: |
|
| Name(s) found: |
UBP23_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 859 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
ubiquitin-dependent protein catabolic process
[IEA]
|
| Molecular Function: |
ubiquitin-specific protease activity
[ISS]
ubiquitin thiolesterase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEVATSSTEI TIQTDRDPSS NNNGSCAVAS STASAVFRKI EFHPARKPFN GFSNGRSDFK 60
61 IETLNPCSSN QRLLSAPSAK KPDSSDLLEH GFEPDLTFSI TFRKIGAGLQ NLGNTCFLNS 120
121 VLQCLTYTEP LAATLQTAAH QKYCHVAGFC ALCAIQKHVR TARQANGRIL APKDLVSNLR 180
181 CISRNFRNCR QEDAHEYMIN LLECMHKCSL PSGVPSESSD AYRRSLVHKI FGGSLRSQVK 240
241 CEQCSHCSNK FDPFLDLSLD ISKADSLQRA LSRFTAVELL DNGAKVYQCE RCKQKVKAKK 300
301 QLTVSKAPYV LTVHLKRFEA HRSEKIDRKV DFTSAIDMKP FVSGPHEGNL KYTLYGVLVH 360
361 YGRSSHSGHY ACFVRTSSGM WYSLDDNRVV QVSEKTVFNQ KAYMLFYVRD RQNAVPKNSV 420
421 PVVKKESFAT NRASLIVASN IKDQVNGSTV IKECGFGALV ANGLAPLKSC GPSTPAVLTQ 480
481 KDLNAKETQN NAISNVEAKE ILETENGSAP VKTCDLAAPT VLVQKDLNTK EIFQKEVPLP 540
541 QANGEGSLVK EDSKAACLIL PEKVSPHLDG SANAQTLVKL PTLGPKAENS VEEKNSLNNL 600
601 NEPANSLKVI NVSVGNPPVE KAVLIDQTMG HHLEESATSI ESLKLTSERE TLTTPKKTRK 660
661 PKTKTLKVEF KFFKLALGLR KKKVQRRERL STTVAGEIIS EELLSKKRVK YQDTSLIAPS 720
721 KMISSSDGAV TSDQQQPVGS SDLSEASQNA KRKRESVLLQ KEAVNILTRG VPETVVAKWD 780
781 EEISASQKRG SKSEGASSIG YVADEWDEEY DRGKKKKIRI KEESYRGPNP FQMLASKRQK 840
841 ETKKKWTQSI TDAKTAYRI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
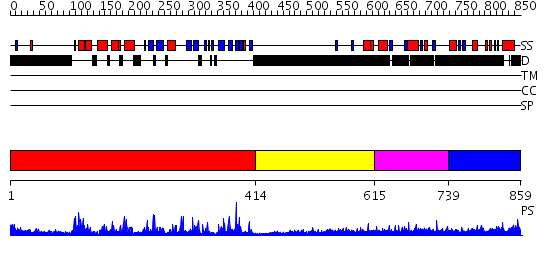
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..413] | 85.045757 | No description for 2gfoA was found. | |
| 2 | View Details | [414..614] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [615..738] | 7.217995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [739..859] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)