













| General Information: |
|
| Name(s) found: |
RH26_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 850 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] ATP-dependent helicase activity [IEA][ISS] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSKFPLGVR FITHSLPCTR LASMNSGALI YSFRSVPVLS KAFPFRLKYI GLGSRVNFST 60
61 RPERSQPEFA RRSGAGGEIR ASKSLIEDEA ELSDWVSDLR TSSLRGKFTS DEDNADPEVV 120
121 RRNVDRDTSR GPRRGREGQS DRFGGAKRGK EGEMDRFGSP NRRRTSGEPA DSFGNKRLGD 180
181 REGSRNGRVQ GKSSESSFRG RSDRNVDSGS SFRGRSDKNV DSGSSFRGRN DRNVDSGSSF 240
241 RGRSDRNVDS GSSFRGRSDR NVDSGSSFRG RNDRNVDSGS SFRGRNDRNV ESGFRREPGS 300
301 ENNRGLGKQT RGLSLEEEDS SDDDENRVGL GNIDDLPSED SSDEDDENDE PLIKKAASAK 360
361 AVQTDKPTGE HVKTSDSYLS KTRFDQFPLS PLSLKAIKDA GFETMTVVQE ATLPIILQGK 420
421 DVLAKAKTGT GKTVAFLLPA IEAVIKSPPA SRDSRQPPII VLVVCPTREL ASQAAAEANT 480
481 LLKYHPSIGV QVVIGGTKLP TEQRRMQTNP CQILVATPGR LKDHIENTSG FATRLMGVKV 540
541 LVLDEADHLL DMGFRRDIER IIAAVPKQRQ TFLFSATVPE EVRQICHVAL KRDHEFINCV 600
601 QEGSGETHQK VTQMYMIASL DRHFSLLHVL LKEHIADNVD YKVIIFCTTA MVTRLVADLL 660
661 SQLSLNVREI HSRKPQSYRT RVSDEFRKSK AIILVTSDVS ARGVDYPDVS LVVQMGLPSD 720
721 REQYIHRLGR TGRKGKEGEG VLLLAPWEEY FMSSVKDLPI TKSPLPPIDP EAVKRVQKGL 780
781 SQVEMKNKEA AYQAWLGYYK SQKMIARDTT RLVELANEFS RSMGLDSPPA IPKNVLGKMG 840
841 LKNVPGLRTK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
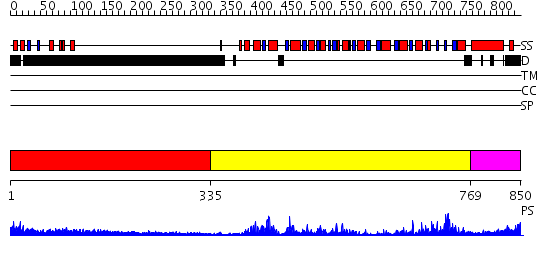
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..334] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [335..768] | 85.045757 | No description for 2db3A was found. | |
| 3 | View Details | [769..850] | 31.69897 | Nucleotide excision repair enzyme UvrB |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.59 |
Source: Reynolds et al. (2008)