













| General Information: |
|
| Name(s) found: |
LISC_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 394 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
metabolic process
[IEA]
lipoic acid biosynthetic process [ISS] lipoate biosynthetic process [IEA] |
| Molecular Function: |
lipoic acid synthase activity
[ISS]
catalytic activity [IEA] lipoate synthase activity [IEA] iron-sulfur cluster binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMHHCSITKP TFSISISTQK LHHHSSKFLN LGFRIRCESG DVSSPLRTKA VSLSSEMEDS 60
61 SSLKKSLMEL EGKKSEPYPG GMPKMGPFTG RDPNVKKPAW LRQKAPQGER FQEVKESLSR 120
121 LNLNTVCEEA QCPNIGECWN GGGDGVATAT IMVLGDTCTR GCRFCAVKTS RNPPPPDPME 180
181 PENTAKAIAS WGVDYIVITS VDRDDIPDGG SGHFAQTVKA MKRHKPDIMI ECLTSDFRGD 240
241 LEAVDTLVHS GLDVFAHNVE TVKRLQRLVR DPRAGYEQSM SVLKHAKISK PGMITKTSIM 300
301 LGLGETDEEL KEAMADLRAI DVDILTLGQY LQPTPLHLTV KEYVTPEKFD FWKTYGESIG 360
361 FRYVASGPLV RSSYRAGELF VKTMVKESYS KSLS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
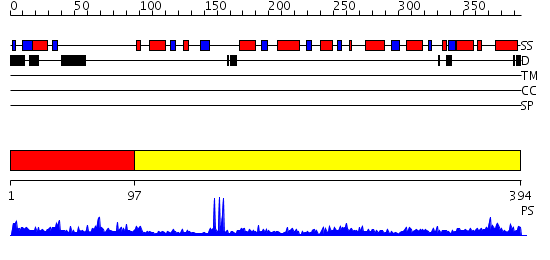
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | 2.754996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [97..394] | 40.39794 | The Crystal Structure of Biotin Synthase, an S-Adenosylmethionine-Dependent Radical Enzyme |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)