













| General Information: |
|
| Name(s) found: |
SUVR2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 717 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
nucleolus [IDA] |
| Biological Process: |
chromatin modification
[IEA]
|
| Molecular Function: |
zinc ion binding
[IEA]
histone-lysine N-methyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPNLHIKKA FMAMRAMGIE DARVKPVLKN LLALYEKNWE LIAEDNYRVL ADAIFDSHED 60
61 QAIQESEEKK ADEVKEDEGC AAEVDRGKKK LHESIEDDED VMAESDRPLK RLRRRGEGGS 120
121 ALASPSLGSP TLEGPSINDE ENAPILLPYH PVPIENDHDA GELILTKVEP ITNMPLSSIP 180
181 DSVDRGDSSM LEIDKSNGHV EEKAGETVST ADGTTNDISP TTVARFSDHK LAATIEEPPA 240
241 LELASSASGE VKINLSFAPA TGGSNPHLPS MEELRRAMEE KCLRSYKILD PNFSVLGFMN 300
301 DICSCYLDLA TNGRDSANQL PKNLPFVTTN IDALKKSAAR MAYTSQASND VVEICSNEHM 360
361 RDAENGAVGD SMALVVVPEC QLSADEWRLI SSVGDISLGK ETVEIPWVNE VNDKVPPVFH 420
421 YIAQSLVYQD AAVKFSLGNI RDDQCCSSCC GDCLAPSMAC RCATAFNGFA YTVDGLLQED 480
481 FLEQCISEAR DPRKQMLLYC KECPLEKAKK EVILEPCKGH LKRKAIKECW SKCGCMKNCG 540
541 NRVVQQGIHN KLQVFFTPNG RGWGLRTLEK LPKGAFVCEL AGEILTIPEL FQRISDRPTS 600
601 PVILDAYWGS EDISGDDKAL SLEGTHYGNI SRFINHRCLD ANLIEIPVHA ETTDSHYYHL 660
661 AFFTTREIDA MEELTWDYGV PFNQDVFPTS PFHCQCGSDF CRVRKQISKG KNVKKRA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
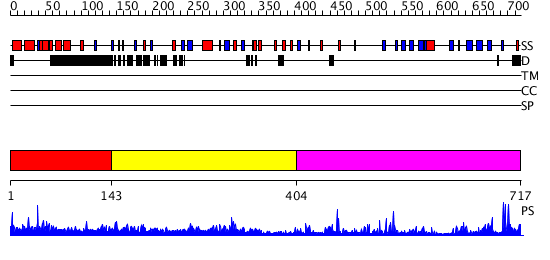
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..142] | 1.172992 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [143..403] | 37.0 | Ankyrin-R | |
| 3 | View Details | [404..717] | 40.154902 | No description for 2o8jA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)