













| General Information: |
|
| Name(s) found: |
RPAP2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 735 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKDNEAIAI NDAVHKLQLY MLENTTDQNQ LFAARKLMSR SDYEDVVTER AIAKLCGYTL 60
61 CQRFLPSDVS RRGKYRISLK DHKVYDLQET SKFCSAGCLI DSKTFSGSLQ EARTLEFDSV 120
121 KLNEILDLFG DSLEVKGSLD VNKDLDLSKL MIKENFGVRG EELSLEKWMG PSNAVEGYVP 180
181 FDRSKSSNDS KATTQSNQEK HEMDFTSTVI MPDVNSVSKL PPQTKQASTV VESVDGKGKT 240
241 VLKEQTVVPP TKKVSRFRRE KEKEKKTFGV DGMGCAQEKT TVLPRKILSF CNEIEKDFKN 300
301 FGFDEMGLAS SAMMSDGYGV EYSVSKQPQC SMEDSLSCKL KGDLQTLDGK NTLSGSSSGS 360
361 NTKGSKTKPE KSRKKIISVE YHANSYEDGE EILAAESYER HKAQDVCSSS EIVTKSCLKI 420
421 SGSKKLSRSV TWADQNDGRG DLCEVRNNDN AAGPSLSSND IEDVNSLSRL ALAEALATAL 480
481 SQAAEAVSSG NSDASDATAK AGIILLPSTH QLDEEVTEEH SEEEMTEEEP TLLKWPNKPG 540
541 IPDSDLFDRD QSWFDGPPEG FNLTLSNFAV MWDSLFGWVS SSSLAYIYGK EESAHEEFLL 600
601 VNGKEYPRRI IMVDGLSSEI KQTIAGCLAR ALPRVVTHLR LPIAISELEK GLGSLLETMS 660
661 LTGAVPSFRV KEWLVIVLLF LDALSVSRIP RIAPYISNRD KILEGSGIGN EEYETMKDIL 720
721 LPLGRVPQFA TRSGA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
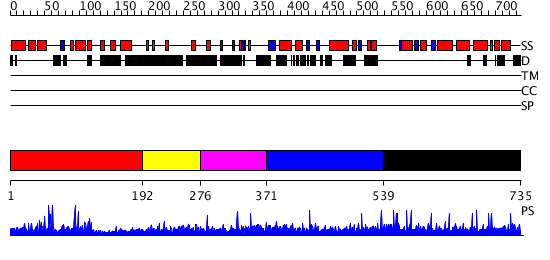
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | 7.041986 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [192..275] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [276..370] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [371..538] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [539..735] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)