













| General Information: |
|
| Name(s) found: |
P2C71_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 447 amino acids |
Gene Ontology: |
|
| Cellular Component: |
protein serine/threonine phosphatase complex
[IEA]
|
| Biological Process: |
protein amino acid dephosphorylation
[IEA]
|
| Molecular Function: |
protein serine/threonine phosphatase activity
[IEA][ISS]
catalytic activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGYMDLALSY SNQMRIVEAP ASGGGLSQNG KFSYGYASSA GKRSSMEDFF ETRIDGIDGE 60
61 IVGLFGVFDG HGGSRAAEYV KRHLFSNLIT HPKFISDTKS AIADAYTHTD SELLKSENSH 120
121 TRDAGSTAST AILVGDRLLV ANVGDSRAVI CRGGNAFAVS RDHKPDQSDE RERIENAGGF 180
181 VMWAGTWRVG GVLAVSRAFG DRLLKQYVVA DPEIQEEKID DSLEFLILAS DGLWDVFSNE 240
241 EAVAVVKEVE DPEESTKKLV GEAIKRGSAD NITCVVVRFL ESKSANNNGS SSSEEANQVP 300
301 TAVRNDSDHK ISAKETNQDH TTVNKDLDRN TDSQSLNQKP IAARSADNSN QKPIATTATG 360
361 HSVSSEQSGL TGEKSQMPIK IRSDSEPKSS AKVPNQTQST VHNDLDSSTA KKPAATEQSG 420
421 STGERNRKPI KVHSDSAARK TTPSIFN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
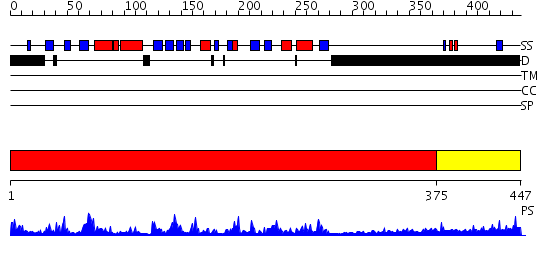
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..374] | 74.522879 | CRYSTAL STRUCTURE OF THE PROTEIN SERINE/THREONINE PHOSPHATASE 2C AT 2 A RESOLUTION | |
| 2 | View Details | [375..447] | 1.099997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)