













| General Information: |
|
| Name(s) found: |
SUSY1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 808 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
sucrose biosynthetic process
[ISS]
response to cold [IEP] response to osmotic stress [IEP] response to flooding [IEP] response to cadmium ion [IEP] |
| Molecular Function: |
UDP-glycosyltransferase activity
[ISS]
sucrose synthase activity [IDA][IGI][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANAERMITR VHSQRERLNE TLVSERNEVL ALLSRVEAKG KGILQQNQII AEFEALPEQT 60
61 RKKLEGGPFF DLLKSTQEAI VLPPWVALAV RPRPGVWEYL RVNLHALVVE ELQPAEFLHF 120
121 KEELVDGVKN GNFTLELDFE PFNASIPRPT LHKYIGNGVD FLNRHLSAKL FHDKESLLPL 180
181 LKFLRLHSHQ GKNLMLSEKI QNLNTLQHTL RKAEEYLAEL KSETLYEEFE AKFEEIGLER 240
241 GWGDNAERVL DMIRLLLDLL EAPDPCTLET FLGRVPMVFN VVILSPHGYF AQDNVLGYPD 300
301 TGGQVVYILD QVRALEIEML QRIKQQGLNI KPRILILTRL LPDAVGTTCG ERLERVYDSE 360
361 YCDILRVPFR TEKGIVRKWI SRFEVWPYLE TYTEDAAVEL SKELNGKPDL IIGNYSDGNL 420
421 VASLLAHKLG VTQCTIAHAL EKTKYPDSDI YWKKLDDKYH FSCQFTADIF AMNHTDFIIT 480
481 STFQEIAGSK ETVGQYESHT AFTLPGLYRV VHGIDVFDPK FNIVSPGADM SIYFPYTEEK 540
541 RRLTKFHSEI EELLYSDVEN KEHLCVLKDK KKPILFTMAR LDRVKNLSGL VEWYGKNTRL 600
601 RELANLVVVG GDRRKESKDN EEKAEMKKMY DLIEEYKLNG QFRWISSQMD RVRNGELYRY 660
661 ICDTKGAFVQ PALYEAFGLT VVEAMTCGLP TFATCKGGPA EIIVHGKSGF HIDPYHGDQA 720
721 ADTLADFFTK CKEDPSHWDE ISKGGLQRIE EKYTWQIYSQ RLLTLTGVYG FWKHVSNLDR 780
781 LEARRYLEMF YALKYRPLAQ AVPLAQDD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
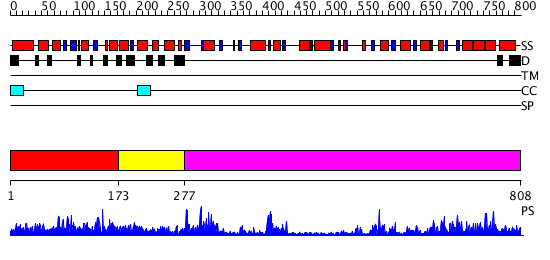
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..172] | 2.024936 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [173..276] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [277..808] | 60.0 | No description for 2r60A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)