













| General Information: |
|
| Name(s) found: |
RUK_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1366 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
embryonic development ending in seed dormancy
[NAS]
phragmoplast assembly [IMP] cytokinesis [IMP] cellularization of endosperm [IMP] |
| Molecular Function: |
kinase activity
[ISS]
microtubule binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNQYHIYEAI GHGKCSTVYK GRKKKTIEYF ACKSVDKSRK NKVLQEVRIL HSLNHPNVLK 60
61 FYAWYETSAH MWLVLEYCVG GDLRTLLQQD CKLPEESIYG LAYDLVIALQ YLHSKGIIYC 120
121 DLKPSNILLD ENGHIKLCDF GLSRKLDDIS KSPSTGKRGT PYYMAPELYE DGGIHSFASD 180
181 LWALGCVLYE CYTGRPPFVA REFTQLVKSI HSDPTPPLPG NASRSFVNLI ESLLIKDPAQ 240
241 RIQWADLCGH AFWKSKINLV QLPTQPAFDD MIGINTKPCL SERNGDRPNK TPPKYREKDR 300
301 KGGSKQNENS IQGSKGHETP IKGTPGGSKA QAKLPSRATE EKHGGRPAAN RQVNILRLSR 360
361 IAKANLQKEN EKENYRRPLP NSNENCAEVK IDNTDMELDF DENNDDEGPD ESEGTENTSC 420
421 AQEERVMSHN ENHRRQRVVS SNVPDENSSA NETPTLGEAR DCHEDQSEPM DMSAAPPSAS 480
481 PQLKTHRGRE TSGVAVNHDS SKAPTSLTDV FWHISDLSVR PVMPSRKSDK EAVHSLSFET 540
541 PQPSDFSKKG KQELEPLNNR IITVLSGSSS GLSEKQNLIR YLETLSTNAD AANILTNGPI 600
601 MLVLVKVLRL SKTPAFRVQI ASLIGLLIRH STSIEDDLAN SGILDSLTNG LRDKHEKVRR 660
661 FSMAALGELL FYISTQNEHK DFKPPESPSK ETRSASGWQV SNALISLVSS VLRKGEDDLT 720
721 QVYALRTIEN ICSQGAYWAT RFSSQDLISN LCYIYKATGK QESMRQTAGS CLVRLARFNP 780
781 PCIQTVVEKL SLKEIASSFV KGSAREQQVC LNLLNMAMIG SHTFTSFGRH LVTLTEEKNL 840
841 FPSLLSIIEQ GTEVLRGKAL LFVAFLCKNS RRWLTNFFCN ARFLPVVDRL AKEKDSYLQQ 900
901 CLEAFVNVIA SIIPGMLDTI TNDIQQLMTG RRHGPVSPLN SRAPVKTNAH LFPVVLHLLG 960
961 SSSFKNKMVT PQVLRQLANL TKLVEASFQG RDDFRVTLLQ VLECITGDAP LVTQNGEIII1020
1021 REILPSLAAI YNGNKDGDAR FLCLKIWFDS LTILLTECTE IEQQISEDLK SISNSHFLPL1080
1081 YPALIQDEDP IPAYAQKLLV MLVEFDYIKI SNLLRHNTVS QCFEFLLGDL SSANVNNVKL1140
1141 CLALASAPEM ESKLLSQLKV VRRIGNLLEF VNAKDMEDFL EPTLSLCRAF LLRSLGNKKG1200
1201 LSSNYTKEPT LLSEASFTFE VDPQECIRDI ADFGSNIGLF LHFAGLDDDT SIAVADIASE1260
1261 CVVLLLKAAS REATTGFLTN LPKITPILDS WRRRKSTELH LLVLKRVLHC LGYACKQYLS1320
1321 QAMILSISGH DVSKINAIVS EMKNSDAAGL NSIASLVAME LQRLPR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
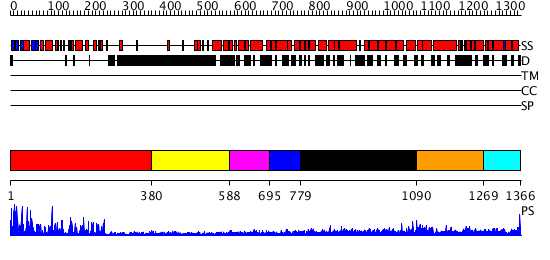
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..379] | 81.30103 | No description for 2r0iA was found. | |
| 2 | View Details | [380..587] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [588..694] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [695..778] | 4.045757 | No description for 2dfsA was found. | |
| 5 | View Details | [779..1089] | 4.0 | Heavy meromyosin subfragment | |
| 6 | View Details | [1090..1268] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1269..1366] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)