













| General Information: |
|
| Name(s) found: |
XLG2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 861 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
response to glucose stimulus
[IGI]
response to abscisic acid stimulus [IGI] G-protein coupled receptor protein signaling pathway [ISS] response to fructose stimulus [IGI] response to mannitol stimulus [IGI] response to sucrose stimulus [IGI] |
| Molecular Function: |
guanyl nucleotide binding
[IEA]
signal transducer activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAVIRKLLP FPSPNPKRDN RESDDDDETS SGYRIEYSFA SEYKGPLIAN VPRALPVEVD 60
61 QIPTALPVSF SSLRSGISYP VAPLVMTKDT KRPPDSGIEK KNGFVDSAAG SSVVLIGRDV 120
121 VSGSSSSSSS KRLDVPEEVK SPADFRLSPS SPLSASAREE DHLDDDRVSD VGPRAVRFVE 180
181 PFQSSECDES SYVSDGESIA ATHRAERKGK RGSCYRCQLG NRFTEKEVCI VCDAKYCFNC 240
241 VRRAMGAMPE GRKCQACIGY RIDESKRASL GKCSRMLKRH LTDSELRQVM NAEITCKANQ 300
301 LPSRLIIVND KPLSEDELYT LQTCPNPPKK LKPGHYWYDK VAGYWGKIGE KPSQIISPNN 360
361 SIGGYISEKV SNGDTEIYIN GREITKPELT MLKWAGVQCE GKPHFWVDSD GSYREEGQKH 420
421 PIGNIWSKKR AKIACAVFSL PVPPASSAVE PYDVPLYEQK MLNKLLLIGS EKGGATTIYK 480
481 QARSLYNVSF SLEDRERIKF IIQTNLYTYL AMVLEAHERF EKEMSNDQSS GNVGDETSAK 540
541 PGNSINPRLK HFSDWVLKEK EDGNLKIFPP SSRENAQTVA DLWRVPAIQA TYKRLRDTLP 600
601 RNAVYFLERI LEISRSEYDP SDMDILQAEG LSSMEGLSCV DFSFPSTSQE ESLESDYQHD 660
661 TDMKYQLIRL NPRSLGENWK LLEMFEDADL VIFCVSLTDY AENIEDGEGN IVNKMLATKQ 720
721 LFENMVTHPS LANKRFLLVL TKFDLLEEKI EEVPLRTCEW FEDFNPLISQ NQTSRHNPPM 780
781 AQRAFHYIGY KFKRLYDSIL EPVNMRGRSF KPKLFVCQVS LESDTVDNAL RYAREILKWH 840
841 VEETSMFQEM STTSIEASSS S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
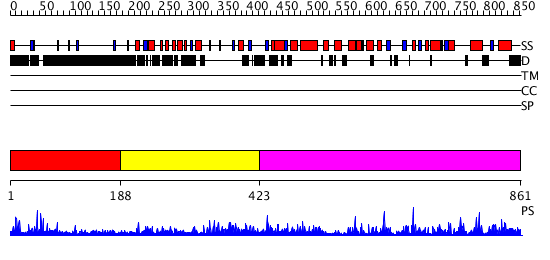
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..187] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [188..422] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [423..861] | 104.0 | No description for 2hlbA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)